Abstract
The microbiome plays a significant role in shaping the health and functioning of the systems it inhabits. The seminal microbiome of stallions has implications for the health of the reproductive tract, sperm quality during preservation and antibiotic use in semen extenders. Diverse bacteria are present on the external genital tract and a mix of commensal microorganisms populates various parts of the reproductive tract, influencing the seminal bacterial content. Other sources of bacteria include the environment, semen collection equipment, and personnel. The bacterial load can adversely affect sperm quality and fertility, particularly in artificial insemination, where semen is extended and stored before use. Antibiotics are frequently used to inhibit bacterial growth, but their effectiveness varies depending on the bacterial strains present. The aim of this study was to assess the bacterial diversity in semen from 37 healthy stallions across three European nations (Germany, Portugal, and Sweden) using 16S sequencing. Semen samples were collected from individual stallions at three AI centers; DNA extraction, sequencing, and bioinformatic analysis were performed. Differences in bacterial diversity among the stallions were seen; although bacterial phyla were shared across the regions, differences were observed at the genus level. Climate, husbandry practices, and individual variability likely contribute to these differences. These findings underscore the importance of tailoring antibiotic strategies for semen preservation based on regional bacterial profiles. The study presents a comprehensive approach to understanding the intricacies of the stallion seminal microbiome and its potential implications for reproductive technologies and animal health.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The microbiome has a marked influence on the reproductive system (Samper 2008; Ortega-Ferrusola et al. 2009; Guimarães et al. 2015; Varela et al. 2018; Al-Kass et al. 2019). This is apparent for the stallion reproductive tract, where the seminal microbiome has considerable implications for the reproductive health of mares bred by the stallions, for retention of sperm quality during semen storage for artificial insemination, and last but not least, for antibiotic usage in semen extenders. Previous studies have examined some of these aspects, ranging from the presence of pathogenic bacteria (Samper 2008), to the impact of bacteria on reproductive technologies (Ortega-Ferrusola et al. 2009; Varela et al. 2018). Other studies described interventions reducing bacterial load (Guimarães et al. 2015; Al-Kass et al. 2019), qualitative and quantitative analysis for bacterial contaminants (Corona and Cherchi 2009); and investigation of the microbial flora in healthy individuals (Rota et al. 2011; Pasing et al. 2013).
Numerous bacteria are constituents of the normal flora on the external genital tract of the stallion, such as Streptococcus dysgalactiae ssp. equisimilis, Bacillus spp., Staphylococcus aureus, Escherichia coli, Streptococcus equi ssp. zooepidemicus, Pseudomonas spp., and Klebsiella spp. (Samper and Tibary 2006). A mixture of commensal bacteria inhabits the surface of the urethral fossa, penis, prepuce, and the urethra of healthy stallions, and are transferred to the semen. Contamination of semen with bacteria from the external genitalia is difficult to avoid when collecting semen with an artificial vagina (Ortega-Ferrusola et al. 2009; Rota et al. 2011). The bacteria present in semen after collection may also originate from the environment (Pickett et al. 1999).
Bacteria in semen can have a deleterious impact on the quality of conserved sperm, affecting their viability and fertility (Aurich and Spergser 2007; Ortega-Ferrusola et al. 2009; Ramires Neto et al. 2015) This is particularly relevant for artificial insemination (AI), where the semen is extended, cooled, and stored for several hours before insemination. Bacterial growth is supported by the nutrients in the semen extender, and bacteria continue to divide at temperatures over 15 °C (Nedwell 1999).
Since bacteria in semen can cause fertility disorders in inseminated mares or may have harmful consequences for semen quality during storage, antibiotics are usually added to inhibit bacterial growth, and the temperature of the semen is reduced. However, the antibiotics added may not inhibit the contaminating bacteria if the incorrect concentration is used or if the microorganisms concerned are not sensitive to their effects (Guimarães et al. 2015). Thus, an understanding of which bacteria are present in stallion semen is necessary to determine the relevant antibiotics to include in semen extenders.
Previous studies on the bacteria present in the semen of stallions without fertility problems were made in Austria (Al-Kass et al. 2019), Germany (Pasing et al. 2013), Italy (Corona and Cherchi 2009; Rota et al. 2011), Portugal (Guimarães et al. 2015), Sweden (Al-Kass et al. 2020) and Spain (Ortega-Ferrusola et al. 2009; Varela et al. 2018; Quiñones-Pérez et al. 2021). The prevalence of the bacteria described varied among studies, perhaps because different techniques were used for bacterial identification but also possibly due to extrinsic factors such as climate, geographic location, husbandry, etc.
Traditionally, bacterial identification is culture-based, which may tend to mask the presence of some bacteria that are difficult to culture (Moretti et al. 2009). Only a few bacteria were identified from stallion semen using culture-based techniques (Corona and Cherchi 2009; Ortega-Ferrusola et al. 2009; Rota et al. 2011; Pasing et al. 2013; Varela et al. 2018; Al-Kass et al. 2019). Recently, studies on the presence of the seminal microbiome in healthy stallions using a non-culture-based method, 16S rRNA sequencing, (Al-Kass et al. 2020; Quiñones-Pérez et al. 2021) revealed many bacterial species, including some that had not been identified previously in culture-based studies.
Research on the human microbiome in the past few years has advanced using next generation sequencing (NGS) (Kuczynski et al. 2012). This method allows the ordering of nucleotides in the DNA to be defined accurately (van der Straaten 2015). The sequencing step is initiated by focusing on genes that code for the 16S (small subunit rRNA gene) of bacteria, using this gene for bacterial taxonomic definitions (Böttger 1989). More than 90% of bacterial genera and 65–83% of bacterial species can be identified in this way (Mignard and Flandrois 2006).
The objective of the present study was to characterize the bacterial species in healthy stallion semen in three European countries using 16S rRNA sequencing, and to evaluate whether climatic factors affect the microbiome.
Materials and methods
Animals and semen collection
Semen samples were obtained by convenience sampling. from 37 stallions at three AI centers in Germany (18 stallions), Portugal (13 stallions), and Sweden (6 stallions), where animals were kept according to national and international regulations. The stallions were free from contagious equine metritis, equine arteritis virus and equine infection anemia virus. In all cases, the stallions were housed in individual boxes with either straw bedding or wood shavings (Portugal), straw or wood chippings (Germany), or wood shavings (Sweden), renewed daily. Access to water was ad libitum. The stallions had access to outside paddocks during the day; in Germany these were sand paddocks, in Portugal grass, and in Sweden sand and grass. The animals were kept according to the European legislation regarding equids. Semen collection with an artificial vagina is considered to be a routine husbandry practice in these countries and therefore did not require ethical permission.
The semen samples were collected during different periods, to fit in with the work-load at the studs: in Portugal from October to May, apart from one in August, in Sweden during September, and in Germany in March. Semen collection was carried out with an artificial vagina (AV) after allowing the stallion to mount a dummy mare. A sterile AV and bottle were used for the collection process, and the penis was not washed before collection at any of the studs. After the collection, the sterile graduated bottle containing the ejaculate was separated from the artificial vagina and taken to the laboratory. All semen collection procedures were performed with sterile equipment and aseptic measures to avoid semen contamination. An aliquot of 1 mL raw ejaculate was stored in liquid nitrogen before transport at − 80 °C to the Clinical Sciences Laboratory at Swedish University of Agricultural Sciences for extraction of bacterial DNA.
Fertility
For the purposes of this study, fertility was arbitrarily categorized arbitrarily as “good” if the stallions had a pregnancy rate per season above 60%, and “low” if the pregnancy rate was below 60% (Table 1).
DNA extraction
The DNA extraction was performed on 10 µL of semen using a QIAamp DNA Mini Kit according to the manufacturer’s instructions for the simultaneous purification of genomic DNA from cells. All samples were centrifuged; the supernatant was removed, and only pelleted cells were used. The purity and concentration of the DNA were tested using a NanoDrop 8000 Spectrophotometer (Thermo Scientific, Waltham (HQ), MA, USA). The DNA purity was considered adequate when the 260/280 ratio was between 1.7 and 1.9, and the concentrations were between 3.87 and 243.83 ng/µL. The DNA samples were stored at − 80 °C until further preparation.
16S rRNA sequencing
A two-step amplification protocol was used to prepare the 16S region of the bacterial DNA content for Illumina sequencing. The primers and cycling protocols are presented in Table 2. The reaction volume of the first step was 21 µL containing 4 µl of sample, 10 µl KAPA HiFi HotStart ReadyMix (Roche), 0.2 µl BSA (Thermo Scientific), 2 µl Primer mix (7.5 µM solution, containing 341 F and 805R, forward and reverse primers), and 4.8 µl ultra-pure water. For the second step, the reaction volume was 20 µL containing 6 µL of the purified DNA template from the first PCR step, 10 µl KAPA HiFi HotStart ReadyMix, and 4 µl of indexing primer mix (i5 and i7 indexing primer, 2.5 µM). Both PCR set-up and bead clean-up were performed in duplicate with Agilent NGS workstation Bravo (Agilent Technologies, USA) in a 96-well plate format. As the samples varied in amounts of bacterial DNA, a test with primers for the first PCR was performed to estimate the appropriate amount for the protocol in each case. The concentrations were estimated with a Qubit 3.0 fluorometer using the High Sensitivity DNA kit. Final bead clean-up was performed after the second PCR using MagSi-NGS prep plus (Tataa). In this step, free primers were removed and the amplicon was purified by binding the DNA to magnetic beads, washing and releasing the DNA in the elution buffer (Qiagen).
16S rRNA analysis
Analysis of 16S rRNA sequencing data was performed using the Nextflow pipeline ampliseq v1.1.2 (https://github.com/nf-core/ampliseq). Briefly, the quality of the sequencing data analysed using FastQC (Andrews 2010), followed by trimming of primer sequences from the reads using cutadapt v2.7 (Martin 2011). Sequencing reads were denoised, dereplicated, and filtered for chimeric sequences using DADA2 (Callahan et al. 2016). Amplicon sequence variants (ASVs) were obtained from the processed sequences and taxonomically classified from phylum to species level after clustering at 99% similarity using the SILVA v132 database (Quast et al. 2013) by applying Naive Bayes classifier (Bolyen et al. 2019). Any ASVs classified as Mitochondria or Chloroplast were removed.
Statistical analysis
For the microbiome data, alpha and beta diversity were calculated using an R package, Phyloseq v1.44.0 (McMurdie and Holmes 2013). Alpha diversity significance for bacterial diversity and richness within the samples was determined using one-way ANOVA with a false discovery rate corrected p-value < 0.05. Species evenness (Pielou) significance was calculated using the Kruskal-Wallis test. Beta diversity significance was determined using overall and pairwise PERMANOVA tests with a Bonferroni corrected p-value < 0.05. LEfSe analysis with the effect size (LDA score) was used to compare the relative abundance between Good fertility (pregnancy rate > 60%) and low fertility stallions from Germany.
Results
The microbiome in semen from stallions in three countries (Germany, Portugal, and Sweden) contained 1,908 identifiable ASVs. Bacterial diversity differed significantly among the different countries (Shannon Index, p = 0.017) (Fig. 1-Left). However, there was no significant difference in richness (observed ASVs, p > 0.05) (Fig. 1-Right) and no significant community evenness (Pielou, p > 0.05) among Germany, Portugal, and Sweden (data not shown).
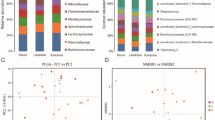
Nineteen bacterial phyla were identified; the ten most frequently seen were Bacteriodetes, Firmicutes, Actinobacteria, Synergistetes, Proteobacteria, Spirochaetes, Patescibacteria, Tenericutes, Fusobacteria, and Epsilonbacteraeota (Fig. 2A). The occurrence of the twenty most abundant bacterial genera is shown in Fig. 2B, with the ten most observed genera being Peptoniphilus, Proteiniphilum, Fastidiosipila, Corynebacterium 1, Petrimonas, Corynebacterium, W5053, Pyramidobacter, Ezakiella, and Lawsonella (Fig. 2B).
Phyla identified in each country are shown in Fig. 3A; the twenty most abundant bacterial genera are shown in Fig. 3B. The dominant phyla in Germany, Portugal, and Sweden were Bacteriodetes, Firmicutes, and Actinobacteria, constituting about 80–95% relative ASV abundance. The three most dominant genera in Germany were Peptoniphilus, Proteiniphilum, and Corynebacterium 1, representing approximately 40% relative ASV abundance. In Portugal and Sweden, Peptoniphilus, Proteiniphilum and Fastidiosipila were dominant, constituting about 70% relative ASV abundance.
The community structure (beta diversity) in stallion semen, analysed using PERMANOVA tests with 10,000 permutations, showed significant differences (p < 0.05) between Germany and the other two countries in Bray-Curtis measurements (Figs. 4, 5 and 6). However, there was no difference (p > 0.05) between Sweden and Portugal. This was also observed with Unifrac and weighted-Unifrac measurements.
Association with fertility
Two genera: Fretibacterium and a previously uncultured genus from Peptostreptococcaceae family appeared in the high fertility stallions to a greater extent than in the lower fertility stallions. The difference was significant with LEfSe analysis, with the effect size (LDA score) 3.9 (p = 0.033) and 3.73 (p = 0.0046), respectively (Fig. 7).
Temperature and precipitation
Mean annual temperatures and rainfall measured at meteorological stations near the studs in Germany, Sweden, and Portugal are shown in Table 3 (https://weather-and-climate.com/). The studs were in temperate regions with a similar maximum temperature in summer, although the number of months in which this temperature was reached was highest in Portugal and least in Sweden. The annual rainfall and number of rainy days was similar between Sweden and Germany, whereas Portugal had almost double the amount of rain but over fewer days. Humidity was similar in the three locations, although the local climate at the stud in Germany was slightly less humid than for the other two sites.
Discussion
The aim of this study was to determine whether there could be differences in the microbiome of semen from stallions kept in different geographical locations. The diversity of bacteria was indeed different between the three countries. The diversity was highest in Germany, then Portugal, and lowest in Sweden. However, the number of stallions from Sweden was much smaller than from the other two countries, due to convenience sampling. There may also have been differences between breeds, but the majority of the stallions in our study were warmbloods, with very few other breeds or types represented. To our knowledge, as yet there is no information about differences in the seminal microbiome among different equine breeds.
Three abundant phyla in our samples in Germany, Portugal, and Sweden, namely Bacteriodetes, Firmicutes, Actinobacteria, were similar to those in Spain (Quiñones-Pérez et al. 2021), in which four abundant phyla in semen samples from 12 stallion were Firmicutes, Bacteriodetes, Actinobacteria, and Proteobacteria. Despite the countries being geographically distant the abundant bacterial phyla were similar. Therefore, these phyla might represent the common bacteria residing in the reproductive tract of healthy stallions in European countries.
Eight abundant genera were Peptoniphilus, Proteiniphilum, Fastidiosipila, Petrimonas Corynebacterium 1, Corynebacterium, Ezakiella, and W5053. However, our results differed from a previous sequencing study in Sweden (Al-Kass et al. 2020) in which ten frequently seen genera were Porphyromonas, Corynebacterium, Finegoldia, Peptoniphilus, Mobiluncus, Chondromyces, Suttonella, Treponema, Acinetobacter, and Campylobacter. Even if the semen samples in the present study were collected from stallions at the same stud in Sweden as in a previous study and contained similar bacterial phyla, they differed at the genus level. The difference in bacterial genera might be due to the semen being collected in different years, or from different individuals, or at different times of the year.
In a previous study based on culturing bacteria from frozen-thawed semen in Portugal, the four genera isolated most frequently were Enterococcus, Staphylococcus, Bacillus, and Corynebacterium (Guimarães et al. 2015). In contrast, in our study using raw semen without antibiotics, the three most abundant genera identified by 16S sequencing were Peptoniphilus, Proteiniphilum, and Fastidiosipila.
The three most abundant genera in Germany from our study were Peptoniphilus, Proteiniphilum, and Corynebacterium 1; these were also different from a previous culture-based study in Germany. In the latter study, the four most frequently cultured genera were Staphylococcus, Streptococcus, Corynebacterium, and Acinetobacter (Pasing et al. 2013). However, the differences in bacteria might be due to the different methodologies used. Interestingly, the genus Corynebacterium was frequently found in stallion semen in both culture- and non-culture based studies.
The ASVs varied between Germany and Portugal, and between Germany and Sweden. A possible explanation for these differences is that many of the bacteria identified are environmental in origin. Therefore, climate and husbandry, as well as season, could be expected to influence the constituents of the microbiome of stallion semen (Al-Kass et al. 2020), at least at the level of genus and species. The stallions were kept in similar husbandry conditions e.g., they were housed individually and had access to paddocks for part of the day. The bedding material was not the same in the three locations, which might affect the bacterial load in the environment, as previously found in studies on cattle bedding (Hogan et al. 1989; Bradley et al. 2018). Pasing et al. (2013) investigated the microorganisms cultured from the genital mucosa and semen of stallions between February and August. They considered that an increase in the occurrence of Staphylococcus vitulinus from swabs of the penile sheath and urethral fossa during May might be due to environmental factors, or to changes in the quality of the bedding, or might be a reflection of an increased workload at the height of the breeding season among the staff caring for the animals. Interestingly, the present study was conducted at the same AI center as the one performed by Pasing et al. (2013),, and both studies may have included some of the same stallions.
The semen samples were collected in a different season in Sweden, which might have been expected to affect the microbial communities present. Most environmental bacteria are mesophilic (Nedwell, 1999) i.e. they do not grow below15°C; the temperature at the time of sampling would have been approximately 0–9 °C in Germany, somewhat lower than in Sweden (9–16 °C) or Portugal (7–19 °C for most of the samples). However, it was generally wetter in Portugal than in Sweden or Germany during the sampling period. Bacterial growth is fastest in conditions of high temperatures and high humidity (Qiu et al. 2022). Survival of bacteria such as Escherichia coli is affected by abiotic factors such as temperature, moisture and solar radiation (Jang et al. 2017), Therefore, one might have expected the conditions to be more favorable to bacterial growth in Portugal than in the other two locations, although this did not appear to be the case from our results. Semen samples from Germany, which experienced the coolest temperatures and an intermediate rainfall during the sampling period, had the greatest diversity of bacteria of the three groups. Therefore, factors other than climate appear to be important in determining the seminal microbiome.
Bacterial diversity might be connected to other factors not mentioned here. In a recent study on the microbial load in bull semen, the length of abstinence between collections (varying from one to three days) was found to influence seminal bacterial load, with the lowest levels in the ejaculates collected after one day´s abstinence (Cojkic et al. 2023). Unfortunately, information on the frequency of semen collection is lacking for the stallions in the present study. Although semen is collected regularly during the breeding season, either three times a week or every day, according to the requirements for semen from particular stallions, the samples from stallions in Portugal were collected when the stallions were presented for a breeding soundness examination rather than as part of a normal routine collection. The samples in Germany were obtained out of season at the start of the period of semen cryopreservation, whereas the end of the previous breeding season would have been August. However, the extra-gonadal sperm reserves had been depleted by semen collection in the week prior to the collections for this study. The period of abstinence in Portugal was likely to have been shorter since the samples were collected during the breeding season. However, a prolonged period of abstinence could explain the findings that the bacterial diversity was higher in the semen samples from Germany. As yet, no studies have been conducted to determine how many semen collections are needed to stabilize the bacterial populations in the reproductive tract.
Such findings could indicate a requirement to investigate the effects of bedding, husbandry, and management practices on microbial load in stallion semen, with a view to reducing antibiotic usage in semen extenders. Current practice is to use commercially available semen extenders containing broad spectrum antibiotics chosen by the manufacturer (Morrell and Wallgren 2014). Knowledge of the specific bacteria present in semen samples from particular locations might enable a more targeted approach to be used, choosing antibiotics that are effective for the bacteria prevalent at that location or finding alternatives (Morrell et al. 2022). Other factors that affect the microbial content of semen include hygiene practices but are not considered here. Such factors include whether or not to wash the penis of the stallion before semen collection, which is a common practice in some countries, or specifically at some studs (Kenney et al. 1975) but not in others (Bowen et al. 1982). In addition, discarding the pre-sperm fraction as in boar semen collection (Goldberg et al. 2013) could have an effect on the bacterial load. Furthermore, allowing the male to make several false mounts before collecting the semen, as in bull semen collection (Şahin et al., 2020) should, theoretically, help to flush bacteria out of the urethra before the ejaculate is collected. However, this practice is seldom used when collecting stallion semen. These factors are beyond the scope of the present study but should be included in extensive studies on controlling the bacterial content of semen. In any case, semen collection and processing should always be carried out with strict attention to hygiene (Althouse 2008).
The two bacteria Fretibacterium (Synergistetes phyla) and the unidentified genus from Peptostreptococcaceae family (Firmicutes phyla) were found abundant in the high fertility group compared to the low fertility group. Usually, an LDA score > 2 is considered as the threshold for a difference to be significant. Fretibacterium was previously isolated from the human oral cavity (Vartoukian et al. 2013). Numerous individuals from the Peptostreptococcaceae family are recognized as common residents of the gastrointestinal system. An unidentified genus of Peptostreptococcaceae was previously observed within domestic cats, constituting a substantial proportion of the fecal microbial population (Bermingham et al. 2018). Five stallions from Germany were found to have this bacterium in their semen samples, suggesting further investigation of the potential connection between cats and stallions at this stud.
There is a connection between the bacteria found in stallion semen and their negative effects on fertility (Varela et al. 2018), as well as their contribution to reduced sperm survival during storage (Ortega-Ferrusola et al. 2009). Therefore, it is important to know which bacteria are present in semen, to devise the best strategy of how to deal with them. It appears that some of the bacteria found in semen may he resistant to the antibiotics currently used in semen extenders (Guimarães et al. 2015). Knowledge of which bacteria are present in semen in a given country or area could help to identify if antibiotics are needed in semen extenders and which antibiotics would be the most suitable to use. The presence of previously unidentified bacteria in the semen samples is interesting, particularly as they are believed to be present in cats, although we do not know if there is a specific connection to cats at the stud farm in Germanys.
Our study population was restricted by necessity, since we could only obtain samples when the studs had access to stallions and had time to collect the semen when it was not needed for commercial AI. Therefore, the number of stallions was different in the different countries, and semen samples were collected at different times of year, with the samples from Sweden being obtained during the autumn. However, the result is striking; the seminal microbiome was different between Germany and Portugal, but was similar between Portugal and Sweden, suggesting that while climate may play a role in determining which organisms are likely to be present, other factors also influence the composition of this community. Further research is required to determine the optimum conditions for husbandry and semen collection routines to optimise the seminal microbiome.
In conclusion, the present results highlight the need for detailed knowledge of the seminal microbiome in stallions from particular geographic areas in order to provide targeted antibiotic usage in semen extenders. Bacteriodetes, Firmicutes, Actinobacteria, Synergistetes, and Proteobacteria were five abundant phyla in the semen samples from all three countries, similar to a previous study on stallion semen from Sweden. However, differences were observed at the genus level between the samples from the three countries, which may be due to differences in climate and husbandry, as well as to individual differences. An abundance of the genus Fretibacterium, as well as an unidentified genus from the Peptostreptococcaceae family, was observed in the high fertility stallions, which has not been documented previously.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
References
Al-Kass Z, Guo Y, Vinnere Pettersson O, Niazi A, Morrell JM (2020) Metagenomic analysis of bacteria in stallion semen. Anim Reprod Sci 221:106568. https://doi.org/10.1016/J.ANIREPROSCI.2020.106568
Al-Kass Z, Spergser J, Aurich C, Kuhl J, Schmidt K, Morrell JM (2019) Effect of presence or absence of antibiotics and use of modified single layer centrifugation on bacteria in pony stallion semen. Reprod Domest Anim 54:342–349.
Althouse GC (2008) Sanitary procedures for the production of Extended Semen. Reprod Domest Anim 43:374–378. https://doi.org/10.1111/j.1439-0531.2008.01187.x
Andrews S (2010) FastQC:A Quality Control tool for High Throughput Sequence Data. In: Babraham Bioinformatics.https://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 30 Jun 2023
Aurich C, Spergser J (2007) Influence of bacteria and gentamicin on cooled-stored stallion spermatozoa. Theriogenology 67:912–918. https://doi.org/10.1016/j.theriogenology.2006.11.004
Bermingham EN, Young W, Butowski CF, Moori CD, Maclean PH, Rosendale D, Cave NJ, Thomas DG (2018) The fecal microbiota in the domestic cat (Felis catus) is influenced by interactions between age and diet; a five year longitudinal study. Front Microbiol 9:365211. https://doi.org/10.3389/FMICB.2018.01231/BIBTEX
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA, Alexander H, Alm EJ, Arumugam M, Asnicar F, Bai Y, Bisanz JF, Bittinger K, Brejnrod A, Brislawn CJ, Brown CT, Callahan BJ, Caraballo-Rodríguez AM, Chase J, Cope EK, Da Silva R, Diener C, Dorrestein PC, Douglas GM, Caporaso JG (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37(8 37):852–857. https://doi.org/10.1038/s41587-019-0209-9
Böttger EC (1989) Rapid determination of bacterial ribosomal RNA sequences by direct sequencing of enzymatically amplified DNA. FEMS Microbiol Lett 65. https://doi.org/10.1016/0378-1097(89)90386-8
Bowen JM, Tobin N, Simpson RB, Ley WB, Ansari MM (1982) Effects of washing on the bacterial flora of the stallion’s penis. J Reprod Fertil Suppl 32:41–45 PMID: 6820065
Bradley AJ, Leach KA, Green MJ, Gibbons J, Ohnstad IC, black DH, Payne, B, Prout VE, Breen JE (2018) The impact of dairy cows’bedding material and its microbial content on the quality and safety of milk –A cross sectional study of UK farms. Int J Food Microbiol 269:36–45. https://doi.org/10.1016/J.IJFOODMICRO.2017.12.022
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods 2016 13:7. https://doi.org/10.1038/nmeth.3869
Cojkic A, Hansson I, Johannisson A, Axnér E, Morrell JM (2023) Single layer centrifugation as a method for bacterial reduction in bull semen for assisted reproduction. Vet Res Commun 2023:1–10. https://doi.org/10.1007/S11259-023-10178-Y
Corona A, Cherchi R (2009) Microbial quality of equine frozen semen. Anim Reprod Sci 115:103–109. https://doi.org/10.1016/j.anireprosci.2008.11.016
Goldberg AMG, Argenti LE, Faccin JE, Linck L, Santi M, Bernardi ML, Cardoso MRL, Wentz I, Bortolozzo FP (2013) Risk factors for bacterial contamination during boar semen collection. Res Vet Sci 95:362–367. https://doi.org/10.1016/j.rvsc.2013.06.022
Guimarães T, Lopes G, Pinto M, Silva E, Miranda C, Correia MJ, Damásio L, Thompson G, Rocha A (2015) Colloid centrifugation of fresh stallion semen before cryopreservation decreased microorganism load of frozen-thawed semen without affecting seminal kinetics. Theriogenology 83:186–191. https://doi.org/10.1016/J.THERIOGENOLOGY.2014.09.003
Hogan JS, Smith KL, Hoblet KH, Todhunter DA, Schoenberger PS, Hueston WD, Pritchar DE, Bowman GL, Heider LE, Brockett BL, Conrad HR (1989) Bacterial counts in bedding materials used on nine commercial dairies. J Dairy Sci 72:250–258. https://doi.org/10.3168/JDS.S0022-0302(89)79103-7
Jang J, Hur H-G, Sadowsky MJ, Byappanahalli MN, Yan T, Ishii S (2017) Environmental Escherichia coli: ecology and public health implications—a review. J Appl Microbiolo 123:570–581
Kenney RM, Bergman RV, Cooper WL, Morse FW (1975) Minimal contamination techniques for breeding mares: techniques and preliminary findings. Proc Am Association Equine Practitioners 21:327–336
Kuczynski J, Lauber CL, Walters WA, Parfrey LW, Clemente JC, Gevers D, Knight R (2012) Experimental and analytical tools for studying the human microbiome. Nat Rev Genet 13.
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12.https://doi.org/10.14806/EJ.17.1.200.
McMurdie PJ, Holmes S (2013) Phyloseq: an R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 8:e61217. https://doi.org/10.1371/JOURNAL.PONE.0061217
Mignard S, Flandrois JP (2006) 16S rRNA sequencing in routine bacterial identification: a 30-month experiment. J Microbiol Methods 67. https://doi.org/10.1016/j.mimet.2006.05.009
Moretti E, Capitani S, Figura N, Pammolli A, Federico MG, Giannerini V, Collodel G (2009) The presence of bacteria species in semen and sperm quality. J Assist Reprod Genet 26:47–56
Morrell JM, Wallgren M (2014) Alternatives to antibiotics in semen extenders: a review. Pathogens 3:943–946
Morrell JM, Malaluang P, Cojkic A, Hansson I (2022) Alternatives to antibiotics in semen extenders used in artificial insemination. Ch in The Global Antimicrobial Resistance Epidemic – Innovative Approaches and Cutting-Edge Solutions, ISBN 978-1-80356-042-7. https://doi.org/10.5772/intechopen.98171
Nedwell DB (1999) Effect of low temperature on microbial growth: lowered affinity for substrates limits growth at low temperature. FEMS Microbiol Ecol 30:101–111. https://doi.org/10.1111/J.1574-6941.1999.TB00639.X
Ortega-Ferrusola C, Gonzaíez-Fernandez L, Muriel A, Macias-Garcia B, Rodriguez-Martinez H, Tapia JA, Alonso JM, Pena FJ (2009) Does the microbial flora in the ejaculate affect the freezeability of stallion sperm? Wiley Online Libr 44:518–522. https://doi.org/10.1111/j.1439-0531.2008.01267.x
Pasing SS, Aurich C, von Lewinski M, Wulf M, Krüger M, Aurich JE (2013) Development of the genital microflora in stallions used for artificial insemination throughout the breeding season. Anim Reprod Sci 139:53–61. https://doi.org/10.1016/j.anireprosci.2013.03.009
Pickett BW, Voss JL, Jones RL (1999) Control of bacteria in stallions and their semen. J Equine Vet Sci 19:424–469. https://doi.org/10.1016/S0737-0806(99)80254-8
Qiu Y, Zhou Y, Chang Y, Liang X, Zhang H, Lin X, Qing K, Zhou X, Luo Z (2022) The effects of Ventilation, Humidity, and temperature on bacterial growth and bacterial genera distribution. Int J Environ Res Public Health 19:15345. https://doi.org/10.3390/ijerph192215345PMID: 36430064; PMCID: PMC9691097
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. https://doi.org/10.1093/NAR/GKS1219
Quiñones-Pérez C, Hidalgo M, Ortiz I, Crespo F, Vega-Pia JL (2021) Characterization of the seminal bacterial microbiome of healthy, fertile stallions using next-generation sequencing. Anim Reprod 18:e20200052. https://doi.org/10.1590/1984-3143-AR2020-0052
Ramires Neto C, Sancler da Silva YFR, Resende HL, Nascimento Guasti P, Montiero GA, Melo Papa P, Delláqua JA, Junior P, Filho JNP, Alvarenga MA, Papa FO (2015) Control methods and evaluation of bacterial growth on fresh and cooled stallion semen. J Equine Vet Sci 35:277–282. https://doi.org/10.1016/j.jevs.2015.01.014
Rota A, Calicchio E, Nardoni S, Fratini F, Ebani VV, Sgorbini M, Panzani D, Camillo F, Mancianti F (2011) Presence and distribution of fungi and bacteria in the reproductive tract of healthy stallions. Theriogenology 76:464–470. https://doi.org/10.1016/j.theriogenology.2011.02.023
Şahin D, Baştan I, Beste Çil, Tekin K, Akçay E, Daşkın A, Stelletta C (2020) The number of false mounting affects the quality of semen in bulls. Livest Stud 60:9–15
Samper JC (2008) Equine breeding management and artificial insemination. Elsevier Health Sciences
Samper JC, Tibary A (2006) Disease transmission in horses. Theriogenology 66:551–559. https://doi.org/10.1016/j.theriogenology.2006.04.019
van der Straaten T (2015) Next-Generation Sequencing:Current Technologies and Applications. Edited by Jianping Xu. ChemMedChem 10:.https://doi.org/10.1002/cmdc.201402456
Varela E, Rey J, Plaza E, Munoz de Propios P, Ortiz-Rodriguez JM, Alvarez M, Anel-Lopez L, Anel l, De paz P, Gil MC, Morrell JM, Ortega-Ferrusola C (2018) How does the microbial load affect the quality of equine cool-stored semen? Theriogenology 114:212–220. https://doi.org/10.1016/j.theriogenology.2018.03.028
Vartoukian SR, Downes J, Palmer RM, Wade WG (2013) Fretibacterium Fastidiosum gen. nov., sp. nov., isolated from the human oral cavity. Int J Syst Evol Microbiol 63:458–463. https://doi.org/10.1099/IJS.0.041038-0
Wu L, Wen C, Qin Y, van Nostrand JD, Ning D, Sun B, Xue K, Liu F, Deng Y, Liang Y, Zhou J (2015) Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol 15:125. https://doi.org/10.1186/s12866-015-0450-4t
Acknowledgements
Special thanks to Katrin Eriksson and Johan Hellander at Menhammar Stuteri, and the staff at the artificial insemination centers who collected the semen samples used in this study.
Funding
Open access funding provided by Swedish University of Agricultural Sciences. Funded by Mahasarakham University and a grant from the Linne and Axel Ericsson´s stipendium fund, SLU. The authors acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. We thank SLU Bioinformatics Infrastructure (SLUBI) for the management and processing of the sequencing data. Support from NBIS (National Bioinformatics Infrastructure Sweden) is gratefully acknowledged. This project was supported by the UMBLA platform at SLU.
Open access funding provided by Swedish University of Agricultural Sciences.
Author information
Authors and Affiliations
Contributions
Material for this study was provided by CN, CA, TG, AR, PM performed the laboratory work, aided by YG; JM designed the study and applied for the funding from SLU; AN was responsible for the bioinformatic analysis and providing the Figures; PM drafted the manuscript which was reviewed by all authors.
Corresponding author
Ethics declarations
Animal ethics
The stallions at the AI stations were not considered to be experimental animals, since they are kept for the specific purpose of semen collection with an artificial vagina; therefore, no ethical permission was required for this project.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Malaluang, P., Niazi, A., Guo, Y. et al. Bacterial diversity in semen from stallions in three European countries evaluated by 16S sequencing. Vet Res Commun 48, 1409–1421 (2024). https://doi.org/10.1007/s11259-024-10321-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11259-024-10321-3