Abstract
Translocation is an important conservation tool for reducing the probability of extinction of threatened plants. It is also becoming an increasingly common management practice, as habitats are destroyed and climate change pushes more plants beyond the limits of their tolerances. Here we outline the case for informing translocations with dedicated genomic data. We begin by describing principles for using genomic and genetic approaches to enhance the efficiency and success of translocation actions. This includes ensuring that translocated populations are adaptively representative, diverse, and composed (to the greatest possible extent) of unrelated individuals. We then use two Australian case studies to illustrate how these principles have been applied in practice and in a resource-efficient way. For Prostanthera densa, we describe how genomic data have quantitatively informed complex decisions, such as whether, and how extensively, to mix individuals from spatially isolated populations in translocated populations. For Fontainea oraria, genomic data have been used during post-translocation monitoring to confirm that newly established populations incorporate and recombine the little diversity that remained in wild individuals. Overall, we illustrate how a simple workflow can support the development and planning of genomic studies and translocation activities in tandem. In order to ensure greater adoption of translocation genomic workflows, funding bodies in charge of biodiversity management and conservation must direct the necessary resources towards them.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Plant translocations transfer plant material into currently depleted populations or sites where populations were known or were assumed to have been found historically, with the aim of re-establishing self-sustainability (Gaywood et al. 2022). Recent attention has been directed to the restoration of populations that are fit and capable of responding to changing selective pressures (Leger et al. 2021). This is particularly relevant within a context of increased activities during the U.N. Decade on Ecosystem Restoration (www.decadeonrestoration.org) aimed at abating the developing climate crisis (Forzieri et al. 2022) and decreasing current extinction rates (Turvey and Crees 2019). Consequently, planning and on-the-ground activities supporting ecological restoration and threatened species translocation projects are increasingly sophisticated and can rely upon a range of science-based criteria to measure success (Commander et al. 2018; Gann et al. 2019).
Quantifying and qualifying the distribution, amount and nature of genetic diversity can influence the establishment of population trajectories, guide corrective actions and prevent catastrophic population declines (Schwartz et al. 2007; Supple and Shapiro 2018; De Woody et al. 2021). Yet, despite broader recognition that gene-level diversity is critical to biodiversity conservation (e.g., the latest Convention on Biological Diversity, www.cbd.int/conferences/2021-2022/cop-15/documents), the use of relevant diversity measures by translocation practitioners can sometimes be overlooked because of the perception that obtaining genetic information can be complex and specialised (Pierson et al. 2016; Silcock et al. 2019). In the absence of relevant genetic information, generalised best-practice guidelines on how to source plant material are often relied upon as a fall-back position (e.g., IUCN/SSC 2013; National Species Reintroduction Forum 2014; Harrison et al. 2021). However, empirical evidence suggests that generalisations around the distribution of genetic diversity can be misleading even among closely related and co-distributed species (Rossetto et al. 2020), and consequently translocations supported by species-specific genetic knowledge are likely to provide improved long-term outcomes (Fremout et al. 2021).
With the advent of restoration and conservation genomics (Breed et al. 2019; Hogg et al. 2022; Theissinger et al. 2023) and the convergence towards representative, replicable and standardised genetic targets and indicators (Frankham 2022; Hoban et al. 2022), innovative toolsets can guide translocation-based activities towards redressed survivorship trajectories (Chen et al. 2022). Technological advancements have been employed to develop all-inclusive and far-reaching management strategies for model species (e.g., Lovell et al. 2021) but arguably, the magnitude of detail provided by such case studies are not indispensable to provide real-time conservation progress across large numbers of threatened species. Broadly applicable, intuitive and resource-efficient genetic workflows that directly guide on-the-ground activities can lead to a wide spectrum of replicable management actions across multiple species (Rossetto et al. 2021). Obtaining taxon-specific genetic and genomic knowledge that supports the translocation of diverse and resilient populations, is conceivably more accessible than on-the-ground practitioners might realise.
Genetic and genomic knowledge is useful for describing the distribution of diversity, prioritising and designing management strategies such as translocations (Mijangos et al. 2015), as well as monitoring the success of these actions (e.g., by assessing temporal variations in genetic indices; Moreno–Mateos et al. 2020). Direct applications include identifying source populations for future translocations and geographic areas that could cause unwanted admixture (Shapcott et al. 2015), and the potential risks associated with outbreeding depression (Kaulfuß and Reisch 2017) and hybrid swamping (Rutherford et al. 2019). Once evolutionarily eligible sources are identified, the number and composition of individuals to be planted can be optimised to ensure that translocated populations are genetically diverse and representative of the adaptive diversity that exists across the selected range of natural sites (Frankham et al. 2017; Faillace et al. 2021). Careful delineation of source material can serve as a form of genetic rescue in highly clonal or inbred populations or, more generally, where the paucity of unrelated mates limits reproductive success or triggers inbreeding depression (Spielman et al. 2004; van Rossum and Raspe 2018; Kardos et al. 2021; Willi et al. 2022). The principles for assembling individuals for translocation are comparable to those for establishing ex-situ plant collections, and the rich literature on this topic (e.g., Marshall and Brown 1975; Schoen and Brown 1993; Griffith and Husby 2010; Hoban and Schlarbaum 2014; Hoban 2019) is of relevance.
An appropriate genetic dataset can be used to estimate measures of diversity under different ‘candidate’ translocation scenarios. Here a candidate design might consist of picking a specific set of cuttings or seedlings from available collections of the target species. Designs that optimize for a specific diversity measure or objective can also be generated, noting that objectives may vary depending on the target species and its management plan. For instance, in circumstances where closely related individuals need to be incorporated into a population (such as a critically endangered species with few individuals left), it might be useful to minimize the average extent of relatedness between spatially proximate individuals, as measured by mean kinship (e.g., Bragg et al. 2020). In other circumstances, we might design a population using individuals that do not share recent ancestry, obviating the need to minimize measurable kinship, but it might nevertheless be useful to maximize other measures such as allelic diversity. For instance, maximizing gene diversity was found to be useful for identifying population designs that captured high levels of allelic variation (Bragg et al. 2021) while being potentially less affected by missing data or rare genotyping errors.
Genetic or genomic datasets can also be used to guide the design of translocated populations with multiple objectives or conflicting priorities. One useful example would be choosing individuals that are genetically diverse as well as enriched for an important property or trait (Bragg et al. 2022). Here the trait could be a measured phenotype indicating climate-readiness or disease resistance, or genomic markers associated with these desirable traits (Bragg et al. 2022). Similar principles have been incorporated into breeding and management of agronomic species (e.g., Schoen and Brown 1993; Meuwissen 1997; Kinghorn 2011; Reeves and Richards 2018; Mascher et al. 2019; Varshney et al. 2021). Another example would be assembling a population that preserves genetic diversity in a threatened species or population, while also trying to manage risks associated with genetic swamping (Bragg et al. 2021). In both examples, there is potential for conflict between the two discrete priorities and multi-objective optimization can be used to explicitly consider translocation designs that represent different points on the trade-off between the objectives. In such cases the goal is to quantify the trade-offs so that management decisions and monitoring practices can be made based on an informed judgement of the importance of the two different objectives.
Finally, post-translocation monitoring will enhance outcomes from translocated populations designed with genetic/genomic datasets in several strategic ways (Rodríguez–Rodríguez et al. 2018). First, monitoring can facilitate multiple ‘corrective’ iterations of population design, especially through vulnerable phases of establishment, where there might be elevated levels of attrition of plants. This might alter population composition and could result in the loss of substantial diversity. In such cases, it is possible to replace the lost plants in a way that is optimized according to an objective (e.g., to minimize mean kinship or maximise disease resistance), but that observes constraints that include the existing plants in the translocated population, and the stocks of replacement plants that are available (e.g., in a nursery; Bragg et al. 2020). Second, over longer time scales, monitoring of reproductive success and other demographic outcomes will help us better identify approaches to population design that are most often linked to success (Van Rossum and Hardy 2022). Subsequently, genotyping of offspring from the translocated population can also enable a better understanding of the heritability of desirable traits, the monitoring of new genetic recombinants or the contribution of adults to the next generation.
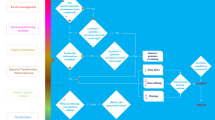
Species-specific genomic datasets obtained from standardised workflows can support various conservation actions including in-situ management prioritisation, the design of multiple translocation strategies, and the establishment of germplasm collections. In this paper, we report two new genomic-based case studies on threatened species, which provide practical guidance towards the development of translocation strategies aimed at minimising extinction risk and maximising evolutionary resilience. The studies support different stages of the translocation process (i.e., planning evolutionary informed translocation scenarios, and monitoring translocation success) and follow a standardised multipurpose workflow as outlined in Box 1. Critical to the workflow are the careful consideration of the sampling strategy (what, why and how many), a standardised process for gathering relevant metadata suitable to multiple downstream processes (and guiding individual labelling), and a standard set of genomic analyses and interpretations.
The Prostanthera densa A.A.Ham. (Villous Mint-Bush, Lamiaceae) example focuses on the optimisation of translocation designs based on inter- and intra-specific levels of genetic differentiation. This species is limited to a few highly localised populations distributed across a restricted range in central New South Wales (Australia; Fig. S1) and is vulnerable to ongoing habitat loss and climate change. An additional complication for P. densa is the frequent confusion between it and another closely related, sympatric and also endangered taxon (P. marifolia R.Br.; Wilson et al. 2012). Prostanthera densa is primarily distinguished from P. marifolia by its growth habit, with a preliminary phylogenetic study using chloroplast and nuclear markers rejecting the interpretation that these differences are due to phenotypic plasticity (Conn et al. 2013).
The Fontainea oraria Jessup and Guymer (Coastal Fontainea, Euphorbiaceae) example focuses on using post-translocation monitoring to assess the success of previously established genetic rescue plantings. This extremely rare species is represented by ten remaining adults and a small number of seedlings within a single coastal population in northern New South Wales (Australia; NSW Department of Planning and Environment 2019). The critically small population exists at four distinct sites of littoral rainforest regrowth within a 600 m radius. An earlier study determined that genetic diversity was unevenly distributed among adults and seedlings, suggesting the over contribution of some genotypes to successive generations (Rossetto et al. 2000). Consequently, a genetic rescue program was initiated to recover viable and self-sustaining Fontainea oraria populations (Department of Environment, Climate Change and Water NSW, 2011). Propagated cuttings from each of the ten adults were planted in a circular design across multiple translocated populations (24 at last count) in order to maximise crossbreeding opportunities and establish new genetic combinations (Brown et al. 2016). These plantings were staggered across years to evaluate planting success. To date, seed and seedlings have been recorded at five translocation sites, and we were able to initiate a conservation genomic study to monitor crossbreeding success.
This study highlights how current genomic tools can support a range of translocation scenarios in a simple, resource-efficient manner and presents novel case studies that are exemplary of this potential.
Material and methods
Target species and sampling strategy
Prostanthera densa (Lamiaceae) is one of 19 taxa listed as Threatened on the Environment Protection and Biodiversity Conservation Act for this large (105 species) and economically valuable genus (Conn et al. 2021). A recovery program including recommendations for translocation was developed (Department of the Environment, Water, Heritage and the Arts 2008), and in its support we conducted a population genomics study that sampled populations of P. densa, P. marifolia and closely related taxa to phylogenetically test the species concept in line with the framework of Rossetto et al. (2021). After testing the species concept for P. densa, we could use the same dataset to assess the extent of clonality, kinship and genetic diversity at the population scale in order to inform management strategies including genetic rescue by way of translocated populations.
One hundred and three specimens of P. densa were sampled, with 98 representing wild-sourced individuals from all known populations plus five ex-situ samples (Table 1). To this we added 48 wild-sourced and five ex-situ P. marifolia individuals originally sourced from the two extant populations. Following the Prostanthera phylogeny of Wilson et al. (2012), we chose six representative taxa to anchor their placement within the phylogenetic framework of the Prostanthera phylogeny (Fig. 1). Within-population sampling for P. densa and P. marifolia varied according to site, including at least six individuals but with the larger sites (Abraham’s Bosom and Manly Dam) sampled more intensively to increase the likelihood of observing accurate amounts of genetic diversity. Additional ex-situ samples were acquired from the Australian Botanic Garden Mount Annan (ABGMA) and the Hunter Region Botanic Gardens (HRBG) to inform the relative value of their current propagation stocks.
a Splitstree network analysis and b SVDquartets phylogenetic coalescent tree generated from single nucleotide polymorphisms (SNPs) data for Prostanthera densa, P. marifolia and closely related Prostanthera. Both Splitstree and phylogeny included 87 genets of P. densa (blue tips labels), 50 non-clonal specimens of P. marifolia (green tip labels) and six related Prostanthera species (black tip labels) as outgroup. Within the network, red branches and stars represent ex-situ individuals from the Hunter Region Botanic Garden (HRBG) and the Australian Botanic Garden Mount Annan (ABGMA). Substructure observed from the genetic data of this study was incorporated into the figure, with populations deriving from a southern clade assigned a “south” label and populations deriving from a northern clade assigned a “north” label. The coalescent tree (> 80% considered as strong branch support), shows that P. densa and P. marifolia form separate well-supported monophyletic clades
While the current distribution of Fontainea oraria (Euphorbiaceae) is no longer subject to clearing, species recovery has been slow due to long maturation times, poor dispersal and recruitment, and limited habitat availability. Its breeding system, mostly unisexual with only rare instances of monoecy (Brown et al. 2016), likely also contributes to its rarity as proximately distributed male and female plants are required for sexual reproduction.
A total of 123 samples were analysed, including the ten original adults and eight wild seedlings, 78 surviving propagated stems from nine translocation sites and 26 seedlings from five of these sites. All propagated plants at the translocation sites were labelled and, given that seedlings tend to germinate under a mother plant, the putative identity of the mother of each seedling was recorded as part of the monitoring. Putative mothers and all planted individuals at each site with seedlings (sites A, C, D, E, J) were genotyped to genetically confirm the parentage of each seedling. Representative stems from more recently planted sites that are yet to produce seedlings (sites B, F, G, N) were analysed to test the accuracy of plant labelling systems during propagation and planting.
DNA extraction and DArTseq
Approximately 5–10 mg plant material was sampled from each individual and sent to DArT Pty Ltd (Canberra, Australia) for DNA extraction and DArT genotype-by-sequencing analyses (referred to as DArTseq). DArTseq is a reduced representation sequencing approach that involves performing a restriction digest of sample DNA, and then high throughput sequencing of the resulting digestion products (Sansaloni et al. 2011). For each study, the resulting single nucleotide polymorphisms (SNPs) dataset was analysed following well-documented in-house procedures as described below (Rossetto et al. 2019; Bragg et al. 2020). The initial raw data was checked for quality using the filtering scripts implemented by the R package RRtools v1.0. To ensure that only the higher quality DArTseq markers were used for analyses, all SNPs with a reproducibility of less than 96% and which had more than 30% missing data were excluded from the dataset, as were linked SNPs.
Test of species concept for P. densa
A phylogenetic tree attempts to estimate the evolutionary history of a set of samples based on inferred common ancestry rather than by observed genetic or phenetic similarities alone. The coalescent-based phylogenetic tool SVDquartets package ver. 1 (Chifman and Kubatko 2014) implemented in the PAUP software v4.0a (Swofford and Sullivan 2012) was used to estimate relationships among populations of P. densa and P. marifolia. This program is designed for SNP data and can analyse datasets with many specimens and genomic sites (Chou et al. 2015), with the multispecies coalescent model being set up with the following parameters: 100,000 quartets and 1000 bootstrap replicates (as a measure of branch support). All results were examined using at least three independent runs for multispecies coalescent analysis by first allocating samples into a group according to their respective population, then allocating all samples into a group according to their species.
Genetic diversity and structure in P. densa
Understanding the extent and distribution of genetic diversity existing in P. densa supports the prioritisation of individuals and populations for conservation actions. To assess genetic similarity among individuals and populations, we performed a Principal Component Analysis (PCA) on the SNPs data using Adegenet 2.1.1 package (Jombart 2008) on R (version 3.3.0, R Core Development Team). Genetic structure was studied by running an admixture analysis using LEA 2.4.0 (Frichot and François 2015). This analysis uses the function snmf (sparse Non-Negative Matrix Factorization algorithms; Frichot et al. 2014) to estimate individual admixture coefficients from large genotypic matrices and evaluate the optimal number of ancestral populations. A measure of fit (i.e., the entropy criterion) is then evaluated between the statistical model and the data to choose the best number of ancestral populations (K) that explain the data. Ten replicates were run for each value of K (up to 8), wherein optimal K was selected by examining the post-stabilisation of the steepest decline in cross-entropy values, with the best replicate of optimal K selected from the lowest minimal cross-entropy values among the 10 replicates. Relevant population genetic diversity measures for each population (i.e., expected heterozygosity (HE), observed heterozygosity (HO) and the inbreeding coefficient (FIS)) were estimated using diveRsity (Keenan et al. 2013) in R.
Designing ex-situ propagation populations of various sizes for P. densa
Small population size and restricted distribution makes species that are threatened by encroaching urbanisation highly susceptible to extinction via stochastic processes. As seed production has not been recorded for P. densa, quick recovery through natural re-establishment or expansion is unlikely. Long-term survival therefore requires the implementation of translocation activities, as suggested in the species’ recovery plan. The development of a translocation plan required a genetic-based understanding of diversity before target plants could be selected from across sites.
To accomplish this, the available SNP dataset was used to provide achievable scenarios for establishing new populations with optimized genomic diversity. As we also identified that P. densa is characterised by two highly distinct genetic groups that lacked admixture among individuals, we provided two distinct scenarios: Scenario 1 targeting maximum diversity within individual genetic groups (in case the two groups are genetically incompatible); Scenario 2 augmenting genetic diversity in the optimisations by incorporating both genetic groups and maximise overall resilience.
Ex-situ propagation populations to prepare for translocation were designed using genomic-based optimisation methods developed by Bragg et al. (2020). Population designs were modelled using the simulated annealing optimisation algorithm (Kirkpatrick et al. 1983), choosing subsets of available plants that maximise genetic distance. The optimisation of individuals used in the scenarios, was based on simulated predictions of genetic diversity remaining after 10 generations of genetic drift, with the Scenario 2 implementing a two-step optimisation analysis. We simulated genetic drift to account for the absence of gene flow between P. densa sites (as these were planned to be disjunct). Each optimised translocation design involved considering propagation populations of different sizes and selecting the appropriate cohort of propagules representing the maximal extent of genetic diversity directly sourced from cuttings of wild individuals.
Relatedness in F. oraria and genetic rescue success
Genetic rescue success within translocated sites was monitored by assessing mating success between translocated adults. This involved measuring relatedness or kinship among all translocated individuals (adults and seedling) via an identity-by-descent analysis of the SNP data that estimates pairwise kinship coefficients between all individuals (using the R package SNPrelate v1.17.1; Zheng and Zheng 2013). The pairwise matrix was drawn using ComplexHeatmap (Gu et al. 2016) in R and was visualised as a network drawn using igraph and visNetwork in R (Csardi 2013; Almende and Thieurmel 2019).
Pairwise kinship coefficients of 0.4 or greater were used to indicate genetic identity (among the propagated individuals in relation to the wild adults) and track the reliability of labelling during propagation. Kinship less than 0.4 was used to study parent–offspring relationships across all individuals in the wild and at the translocation sites. Parentage for each seedling could be determined by observing the first and second highest kinship values between the seedling and any adult. The identity of a parent was verified against the observed maternal parent, except where the observed identity was uncertain (i.e., identity of propagated plant not matching its genetically determined identity). Parent combinations were tabulated based on the parentage data across all seedlings to study the mating patterns between adults in the wild and at translocation sites.
The original genetic rescue program developed for F. oraria aimed to establish multiple new reproductive populations producing seedlings that are representative of the diversity within the parental generation and include admixture events that are currently unattainable in the wild. As a result, we also used the SNP dataset to assess the genetic representativeness of the translocated seedlings. To achieve this, we estimated the total number of SNPs that were ‘common’ (minor allele frequency > 3%) and calculated the proportion of these SNPs that were polymorphic for the seedling cohort and adults at the wild and translocation sites.
Results
Prostanthera densa—Translocation scenarios to improve the long-term viability of a threatened species
We obtained 46,000 genome-wide markers (SNPs) for 163 samples representing P. densa, P. marifolia and related species of Prostanthera. Coalescent analysis and the Splitstree network inferred a topology that consistently indicated that P. densa and P. marifolia formed separate clades, supporting their distinction. Although both formed the closest relationship with each other respective of the outgroup, the two P. marifolia sites formed a clade sister to a clade of all seven P. densa sites in the phylogeny (Fig. 1b). Whether samples were allocated into seven groups respective of their site membership, or within two groups respective of their species membership, the coalescent analysis consistently provided a high number of compatible quartets, suggesting a relatively low amount of incomplete lineage sorting between populations as well as between the species. The SDVquartets analysis at the population scale retrieved a total weight of incompatible quartets = 7,788 (7.79%) and a total weight of compatible quartets = 92,212 (92.21%); when populations were grouped into their hypothesised species, the analysis retrieved a total weight of incompatible quartets = 7.6544 (21.87%), and total weight of compatible quartets = 27.3456 (78.13%). Nonetheless, the connection point of the network between populations of both species was broader than the stems supporting each site, suggesting that reticulation may have occurred (Fig. 1a).
The substructure within P. densa corresponded with geographic separation of sites. In the Splitstree network and phylogeny (Fig. 1), sites further north of Helensburgh (i.e., Royal NP, Bass Flinders, Karrara hill, and Gaan Gaan hill) were recovered as one clade and those in the south (i.e., Helensburgh, Abraham's Bosom, Honeymoon) formed a second clade (hereafter referred to as the northern and southern groups, see Fig. 1). Output from LEA likewise showed this partition, returning two ancestral populations (K = 2) that, with the exception of the Helensburgh population, corresponded to the northern and southern groups (Fig. 2). Geographically proximal sites were more genetically similar (Karrara hill to Gaan Gaan hill, Royal NP Marley to Bass Flinders, Abrahams Bosom to Honeymoon Bay; Fig. 1a, Fig. 2b), with higher pairwise Fst estimates detected between more geographically distant sites (Fig. S3). Despite a latitudinal distribution of genetic divergence, a comparison between genetic and geographic diversity measures did not produce a simple isolation by distance trend (Fig. S3). Finally, the geographically intermediate Helensburgh site displayed a mixture of both ancestral populations (although proportionally more similar to the northern group; Fig. 1a). However, when additional ancestral populations (> K = 3) were requested in the LEA analysis, Helensburgh formed a distinct genetic signature from other sites (Fig. S2) alluding to a possible hybrid-origin scenario.
Genomic diversity outputs for Prostanthera densa showing two strongly distinct genetic clusters (identified as red and blue) in the plots. a Population structure output based on averaged ancestral assignment of K = 2 across northern (Gaan Gaan hill, Karrara hill, Royal NP and Marley) and southern (Helensburgh, Abrahams Bosom and Honeymoon Bay) sites of Prostanthera densa in New South Wales. This optimal number of ancestral populations represented the termination of the steepest decline in associated cross-entropy values. b Principal Component Analysis (PCA) of SNP data from individuals of P. densa. Both analyses identified two strongly distinct genetic clusters
Clonality was detected within multiple sites (Fig. S4): a genet of seven ramets at Gaan Gaan hill, a genet of four ramets at Bass Flinders, three genets consisting of two ramets each at Abrahams Bosom and a genet of two ramets at Honeymoon Bay. The genet of multiple ramets from Gaan Gaan hill shared kin with another genet at the site and both are represented ex-situ, respectively at HRBG and the ABGMA (Fig. 1a).
With the necessary understanding of the distribution of genetic diversity in P. densa, we could provide genomically informed solutions for establishing new populations that removed clonality, reduced kinship and maximised genetic diversity across two scenarios (Fig. 3). For Scenario 1, which separately targeted maximum diversity for northern and southern groups, the analysis identified 10 plants for the northern group and 18 for the southern group as key individuals needed to establish a new population representing 95% of the groups’ genetic diversity. Scenario 2, aimed at combining groups to obtain a population representing greater than 95% of diversity, required five individuals from each of the northern group and the southern groups. While optimised populations from Scenario 1 only respectively captured 0.51 and 0.55 of the diversity measured across the whole species, optimised populations from Scenario 2 captured between 0.96 and 0.98 of alleles (Fig. 3).
Modelling the proportion of alleles captured for optimised translocated Prostanthera densa sites of various sizes (number of genets). Two solutions are provided: one sourcing either of the two single genetically distinct groups (northern group as red circles, southern group as blue circles), the second (northern group as red stars, southern group as blue stars) sourcing the same groups but including genetic supplementation from the other (north or south) group. The second solution involves first optimising targeted individuals from the main genetic group to identify the most optimal size for propagation (blue and red dots; 10 individuals for north and 18 individuals for south captures greater than 95% of diversity in each group:). Following the preliminary optimisation, a second optimisation analysis used the second genetic group to assess improvements to genetic diversity. This two-step analysis resulted in all propagation sites capturing greater than 95% of species’ diversity (dashed line). See the Supplementary methods for additional details
Fontainea oraria—post-translocation monitoring of a genetic rescue project
A high-quality genome scan based on 11,817 genome-wide SNPs was generated for 123 F. oraria samples. The kinship analysis of the dataset identified relatedness and parentage across wild and translocated sites and matched the identity of each propagated stem to a wild adult. There were 14 incidences where the labelled identity of a propagated stem was a mismatch with the plant’s true identity or where individuals labelled as ‘unknown’ could be identified.
The SNP dataset determined the parentage of all seedlings and revealed that all translocated F. oraria adults contributed to the next generation, with admixture within and between sites producing novel parental combinations (Fig. 4; Table S1). Parentage of almost all seedlings could be traced back to adults within the same translocation site with a few exceptions, such as a site A seedling resulting from 324 × 401 mating where 324 was absent at the site, a site E seedling originating from 404 × 420 where 404 was absent, and two site J seedlings originating from 401 × 420 where 401 was absent. In comparison, within the wild population only three adults contributed to the seedling cohort (mostly 420 × 421 matings).
Network visualising pairwise kinship (> 0.05) between all known genets of Fontainea oraria including all adults and seedlings (full pairwise kinship is plotted as a heatmap in Fig. S5). Each coloured circle (or a node) represents an individual and the colour of the node indicates whether an individual is an adult (black) or seedling at wild (yellow) or translocation (orange) site. A line that links the coloured circle indicates there is kinship (> 0.05) between pairs of individuals. The level of kinship is not indicated in the plot hence the length of the line is meaningless. However, nodes that are surrounded by multiple links indicates shared inheritance
We did not detect high levels of kinship (> 0.4) between parents and seedlings suggesting that self-fertilisation is uncommon. Six of the original adults are closely related (likely resulting from biparental inbreeding in small remnant sub-populations) yet only five of 26 translocation seedlings revealed inbreeding, highlighting the power of well-planned genetic rescue strategies even in cases of extreme rarity. Overall allelic diversity within the 26 seedlings at the translocation sites was considerably higher (98% of overall diversity) than within eight wild seedlings (68%) and comparable to the diversity found in the wild adults (Fig. 5).
Proportion of common alleles captured by Fontainea oraria across wild and translocation sites and across various cohorts (all wild seedlings, all wild adults, and all seedlings that grew at translocation sites). The proportion of common alleles captured at translocated sites are shown as orange circles and the proportion of common alleles captured by all wild seedlings is shown as a yellow circle, by all wild adults is shown as a black circle and by all translocated seedlings orange star
Discussion
We demonstrated how a standardised genomic workflow (Box 1) can address translocation-based questions for any target species to inform a range of admixture-based translocation scenarios (including the resolution of taxonomic issues, Prostanthera densa), and post-translocation monitoring (to assess the success of genetic rescue, Fontainea oraria). We advocate that in addition to supporting translocation management, the knowledge gathered by this approach is equally valuable for understanding the ecology and evolutionary history of the target species.
Finalising a suitable propagation strategy from genetic rescue scenarios for Prostanthera densa was dependent on a better understanding of taxonomic boundaries and of the distribution of diversity within the remaining, geographically disjunct populations. A single, carefully planned sampling and analytical workflow including outgroups and co-distributed taxa determined that P. densa and P. marifolia are distinct species with separate conservation priorities. Translocation planning could therefore exclude P. marifolia and focus on complex between-group patterns (i.e., two ancestral populations corresponding to geographically separate northern and southern groups). Interestingly, phylogenetic and phenotypic appraisal revealed that admixture events may have occurred at the P. densa Helensburgh site (Fig. 1). Such findings recommend the exclusion of Helensburgh from the translocation scenarios presented, but also support the development of alternative local and all-inclusive translocation options (Scenarios 1 and 2 respectively).
The same sampling and analytical workflow provided a quantification of evolutionary resilience through measures of clonality, kinship and genetic diversity at the population scale. The presence of higher inbreeding levels within the most geographically isolated populations highlights how reduced gene flow can impact on population-level diversity, irrespective of size. An additional factor in guiding an optimal admixture strategy for translocated populations, was the detection of clonality across numerous populations of P. densa (Fig. S4). This was unexpected as the species is incapable of layering due to its typically upright habit lacking rhizomes or extended branches. Ascertaining the possible impact of apomixis will require dedicated research in the future.
Once the various translocation strategies and scenarios are established, post-translocation monitoring can be used to assess if the planned genetic rescue objectives have been realised and to monitor losses / gains of genotypes through time, as exemplified by the Fontainea oraria example. The original translocation planting design for F. oraria aimed at maximising crossbreeding between genetically and geographically distinct individuals that would otherwise not come into contact in the wild. Post-translocation monitoring shows that genetic rescue was effective, with most of the resulting seedlings originating from crosses between unrelated adults, and with high genetic diversity and novel crosses being generated. The fact that none of the seedlings resulted from self-fertilisation also suggests that the species is a preferential outcrosser. Furthermore, while the mode of seed and pollen dispersal in the wild is unclear (Brunton et al. 2022) the detection of seedlings originating from between-site crosses, suggests that in the right conditions, gene flow can occur across physically separated areas. The monitoring dataset also provided new understanding of the level of relatedness among the wild individuals (Fig. 4, Table S1), supporting the previous suggestion that some of these originated from a shared maternal parent (Rossetto et al. 2000) and validating the need for establishing multiple translocated populations with a genetic rescue focus.
The F. oraria example also identified some mismatches between genetically determined identities and field labels on some translocated individuals. This highlights how a simple genetic study can improve ex-situ resource management, as also demonstrated by the realisation that the ex-situ P. densa collection at the HRBG is all derived from a single genet from Gaan Gaan hill (Supp Fig. 3). Accurate record-keeping is vital for ensuring translocation success (Commander et al. 2018), and ongoing genetic screening is recommended to mitigate mislabelling issues that could occur during the propagation and establishment of translocated plants. Finally, the use of genetically unsuitable ‘leftover’ propagules in additional plantings should be avoided as increased biparental inbreeding or selfing can lead to genetically duplicate siblings (Lu-Irving et al. 2023) and diminish the overall evolutionary resilience of restored populations.
Conclusion
Broadly applicable and resource-efficient genetic workflows that directly guide on-the-ground activities are now accessible and can lead to a wide spectrum of replicable management actions across multiple species (Rossetto et al. 2021; Hogg et al. 2022; Shaffer et al. 2022). These workflows, the product of many years of accumulated knowledge and expertise, are now increasingly accessible and consequently conservation managers should consider from inception how these tools can vastly improve the long-term viability and quality of their translocation projects. However, for these workflows to be more widely adopted it is also essential that the funding bodies in charge of biodiversity management and conservation consistently include them within their granting opportunities.
Increased awareness of the potential of genetics and genomics in translocation can help the conservation community to achieve their goals. The case studies presented relied on single sampling and sequencing events to directly guide suitable translocation strategies, while also revealing more about a species’ ecology and evolutionary history. Leaf tissue needed to generate genetic data for the relevant analyses can often be obtained from one broader sampling event which can be co-ordinated with regular surveys or monitoring actions (therefore further rationalising resources). Sequencing and interpretation of genome-wide SNPs scans were valued at approximately AUD 15,000 (including labour), and a practical report was produced within three months from the completion of field sampling. The efficiency of the process can be further enhanced by considering the economy of scale arising from the simultaneous collection of multiple species (including pre-emptive actions preceding the finalisation of genomic funding). As genomic datasets derived from threatened species are steadily growing, the conservation community will invariably be in a better position to identify threats, interpret patterns, and develop suitable management activities with more predictable outcomes.
Data availability
The data are made available through Dryad (details to be attached on manuscript finalisation).
References
Almende BV, Thieurmel B and Robert T (2019) Package ‘visnetwork’. Network Visualization Using ‘vis. js’ Library 2.
Bragg JG, Cuneo P, Sherieff A, Rossetto M (2020) Optimizing the genetic composition of a translocation population: incorporating constraints and conflicting objectives. Mol Ecol Resour 20:54–65. https://doi.org/10.1111/1755-0998.13074
Bragg JG, Yap JYS, Wilson TC, Lee E, Rossetto M (2021) Conserving the genetic diversity of condemned populations: optimizing collections and translocation. Evol Appl 14:1225–1238. https://doi.org/10.1111/eva.13192
Bragg JG, van der Merwe M, Yap JYS, Borevitz J, Rossetto M (2022) Plant collections for conservation and restoration can they be adapted and adaptable. Mol Ecol Resour 22:2171–2182. https://doi.org/10.1111/1755-0998.13605
Breed MF, Harrison PA, Blyth C, Byrne M, Gaget V, Gellie NJ, Groom SV, Hodgson R, Mills JG, Prowse TA, Steane DA (2019) The potential of genomics for restoring ecosystems and biodiversity. Nat Rev Genet 20:615–628. https://doi.org/10.1038/s41576-019-0152-0
Brown D, James R, McKinley A (2016) Recovery of the Coastal Fontainea and investigation into temporal monoecy. J Aust Netw Plant Conserv 25:11–13. https://doi.org/10.3316/informit.931945134210067
Brunton AJ, Lamont RW, Conroy GC, Yap JS, Rossetto M, Taylor-Brown A, Maggia L, Reddell PW, Ogbourne SM (2022) Phylogenetic Reconstruction of the Rainforest Lineage Fontainea Heckel (Euphorbiaceae) Based on Chloroplast DNA Sequences and Reduced-Representation SNP Markers. Diversity 14:725. https://doi.org/10.3390/d14090725
Chen Z, Grossfurthner L, Loxterman JL, Masingale J, Richardson BA, Seaborn T, Smith B, Waits LP, Narum SR (2022) Applying genomics in assisted migration under climate change: Framework, empirical applications, and case studies. Evol Appl 15:3–21. https://doi.org/10.1111/eva.13335
Chifman J, Kubatko L (2014) Quartet inference from SNP data under the coalescent model. Bioinformatics 30:3317–3324. https://doi.org/10.1093/bioinformatics/btu530
Chou J, Gupta A, Yaduvanshi S, Davidson R, Nute M, Mirarab S, Warnow T (2015) A comparative study of SVDquartets and other coalescent-based species tree estimation methods. BMC Genomics 16:S2. https://doi.org/10.1186/1471-2164-16-S10-S2
Commander LE, Coates DJ, Broadhurst L, Offord CA, Makinson RO, Matthes M (2018) Guidelines for the translocation of threatened plants in Australia, 3rd edn. Australian Network for Plant Conservation, Canberra
Conn BJ, Wilson TC, Henwood MJ, Proft KM (2013) Circumscription and phylogenetic relationships of Prostanthera densa and P. marifolia (Lamiaceae). Telopea 15:149–164
Conn BJ, Henwood MJ, Proft KM, Scott JA, Wilson TC, Howes RS (2021) An integrative taxonomic approach resolves the Prostanthera lasianthos (Lamiaceae) species complex. Aust Syst Bot 34:438–476. https://doi.org/10.1071/SB20023
Csardi MG (2013) Package ‘igraph’. Last accessed 3, 2013.
De Woody JA, Harder AM, Mathur S, Willoughby JR (2021) The long-standing significance of genetic diversity in conservation. Mol Ecol 30:4147–4154. https://doi.org/10.1111/mec.16051
Department of Environment, Climate Change and Water NSW, 2011, Management Plan for Coastal Fontainea Fontainea oraria, Department of Environment, Climate Change and Water NSW, Sydney. https://www.environment.nsw.gov.au/resources/threatenedspecies/20110164MgtPlanCoastal.pdf
Department of the Environment, Water, Heritage and the Arts (2008). Approved Conservation Advice for Prostanthera densa (Villous Mintbush). Canberra: Department of the Environment, Water, Heritage and the Arts. http://www.environment.gov.au/biodiversity/threatened/species/pubs/12233-conservation-advice.pdf. In effect under the EPBC Act from 03-Jul-2008.
Faillace CA, Sentis A, Montoya JM (2021) Eco-evolutionary consequences of habitat warming and fragmentation in communities. Biol Rev 96:1933–1950. https://doi.org/10.1111/brv.12732
Forzieri G, Dakos V, McDowell NG, Ramdane A, Cescatti A (2022) Emerging signals of declining forest resilience under climate change. Nature 608:534–539. https://doi.org/10.1038/s41586-022-04959-9
Frankham R (2022) Evaluation of proposed genetic goals and targets for the convention on biological diversity. Conserv Genet 23:865–870. https://doi.org/10.1007/s10592-022-01459-1
Frankham R, Ballou JD, Ralls K, Eldridge M, Dudash MR, Fenster CB, Lacy RC, Sunnucks P (2017) Genetic management of fragmented animal and plant populations. Oxford University Press, Oxford. https://doi.org/10.1093/oso/9780198783398.001.0001
Fremout T, Thomas E, Bocanegra-Gonzalez KT, Aguirre-Morales CA, Morillo-Paz AT, Atkinson R, Kettle C, González-M R, Alcazar-Caicedo C, Gonzalez MA, Gil-Tobon C (2021) Dynamic seed zones to guide climate-smart seed sourcing for tropical dry forest restoration in Colombia. Forest Ecol Manag 490:119127. https://doi.org/10.1016/j.foreco.2021.119127
Frichot E, François O (2015) LEA: an R package for landscape and ecological association studies. Methods Ecol Evol 6:925–929. https://doi.org/10.1111/2041-210X.12382
Frichot E, Mathieu F, Trouillon T, Bouchard G, François O (2014) Fast and efficient estimation of individual ancestry coefficients. Genetics 196:973–983. https://doi.org/10.1534/genetics.113.160572
Gann GD, McDonald T, Walder B, Aronson J, Nelson CR, Jonson J et al (2019) International principles and standards for the practice of ecological restoration. Restor Ecol 27:S1–S46. https://doi.org/10.1111/rec.13035
Gaywood MJ, Ewen JG, Hollingsworth PM, Moehrenschlager A (Eds.) (2022) Conservation translocations. Cambridge University Press.
Griffith P, Husby C (2010) The price of conservation: measuring the mission and its cost. Bioscience 7:12–14. https://doi.org/10.1093/biosci/biac007
Gu Z, Eils R, Schlesner M (2016) Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. https://doi.org/10.1093/bioinformatics/btw313
Harrison PA, Breed MF, Broadhurst L, Byrne M, Commander L, Coates D, Krauss S, van der Merwe M, Miller M, Rossetto M (2021) FloraBank guidelines module 5: seed sourcing. In: Florabank Guidelines: best practice guidelines for native seed collection and use, Florabank Consortium, Australia
Hoban S (2019) New guidance for ex-situ gene conservation: sampling realistic population systems and accounting for collection attrition. Biol Cons 235:199–208. https://doi.org/10.1016/j.biocon.2019.04.013
Hoban S, Schlarbaum S (2014) Optimal sampling of seeds from plant populations for ex-situ conservation of genetic biodiversity, considering realistic population structure. Biol Cons 177:90–99. https://doi.org/10.1016/j.biocon.2014.06.014
Hoban S, Archer FI, Bertola LD, Bragg JG, Breed MF, Bruford MW, Coleman MA, Ekblom R, Funk WC, Grueber CE, Hand BK (2022) Global genetic diversity status and trends: towards a suite of essential biodiversity variables (EBVs) for genetic composition. Biol Rev 4:1511–1538. https://doi.org/10.1111/brv.12852
Hogg CJ, Ottewell K, Latch P, Rossetto M, Biggs J, Gilbert A, Richmond S, Belov K (2022) Threatened Species Initiative—empowering conservation action using genomic resources. Proc Natl Acad Sci 119:e2115643118. https://doi.org/10.1073/pnas.2115643118
IUCN/SSC (2013). Guidelines for Reintroductions and Other Conservation Translocations. Version 1.0. Gland, Switzerland: IUCN Species Survival Commission, viiii + 57 pp.
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. https://doi.org/10.1093/bioinformatics/btn129
Kardos M, Armstrong EE, Fitzpatrick SW, Hauser S, Hedrick PW, Miller JM, Tallmon DA, Funk WC (2021) The crucial role of genome-wide genetic variation in conservation. Proc Natl Acad Sci 118:e2104642118. https://doi.org/10.1073/pnas.2104642118
Kaulfuß F, Reisch C (2017) Reintroduction of the endangered and endemic plant species Cochlearia bavarica—Implications from conservation genetics. Ecol Evol 7:11100–11112. https://doi.org/10.1002/ece3.3596
Keenan K, McGinnity P, Cross TF, Crozier WW, Prodöhl PA (2013) diveRsity: An R package for the estimation and exploration of population genetics parameters and their associated errors. Methods Ecol Evol 4:782–788. https://doi.org/10.1111/2041-210X.12067
Kinghorn BP (2011) An algorithm for efficient constrained mate selection. Genet Sel Evol 43:4. https://doi.org/10.1186/1297-9686-43-4
Kirkpatrick S, Gelatt CD, Vecchi MP (1983) Optimization by simulated annealing. Science 220:671–680. https://doi.org/10.1126/science.220.4598.671
Leger EA, Agneray AC, Baughman OW, Brummer EC, Erickson TE, Hufford KM, Kettenring KM (2021) Integrating evolutionary potential and ecological function into agricultural seed production to meet demands for the decade of restoration. Restor Ecol. https://doi.org/10.1111/rec.13543
Lovell JT, MacQueen AH, Mamidi S, Bonnette J, Jenkins J, Napier JD, Sreedasyam A, Healey A, Session A, Shu S, Barry K (2021) Genomic mechanisms of climate adaptation in polyploid bioenergy switchgrass. Nature 590:438–444. https://doi.org/10.1038/s41586-020-03127-1
Lu-Irving P, Bragg JG, Rossetto M, King K, O’Brien M, van der Merwe MM (2023) Capturing genetic diversity in seed collections: an empirical study of two congeners with contrasting mating systems. Plants 12(3):522
Marshall DR, Brown AHD (1975) Optimum sampling strategies in genetic conservation. In: Frankel OH, Hawkes JG (eds) Crop genetic resources for today and tomorrow. Cambridge University Press, Cambridge
Mascher M, Schreiber M, Scholz U et al (2019) Genebank genomics bridges the gap between the conservation of crop diversity and plant breeding. Nat Genet 51:1076–1081. https://doi.org/10.1038/s41588-019-0443-6
Meuwissen THE (1997) Maximizing the response of selection with a predefined rate of inbreeding. J Anim Sci 75:934–940. https://doi.org/10.2527/1997.754934x
Mijangos JL, Pacioni C, Spencer PBS, Craig MD (2015) Contribution of genetics to ecological restoration. Mol Ecol 24:22–37. https://doi.org/10.1111/mec.12995
Moreno-Mateos D, Alberdi A, Morriën E, van der Putten WH, Rodríguez-Uña A, Montoya D (2020) The long-term restoration of ecosystem complexity. Nat Ecol Evol 4:676–685. https://doi.org/10.1038/s41559-020-1154-1
National Species Reintroduction Forum (2014) The Scottish Code for Conservation Translocations. Scottish Natural Heritage. https://www.nature.scot/professional-advice/protected-areas-and-species/protected-species/reintroducing-native-species
NSW Department of Planning and Environment (2019) Coastal Fontainea. Available at: https://www.environment.nsw.gov.au/threatenedspeciesapp/profile.aspx?id=10334. Accessed 12 Oct 2022
Pierson JC, Coates DJ, Oostermeijer JGB, Beissinger SR, Bragg JG, Sunnucks P, Schumaker NH, Young AG (2016) Genetic factors in threatened species recovery plans on three continents. Front Ecol Environ 14:433–440. https://doi.org/10.1002/fee.1323
Reeves PA, Richards CM (2018) Biases induced by using geography and environment to guide ex-situ conservation. Conserv Genet 19:1281–1293. https://doi.org/10.1007/s10592-018-1098-z
Rodríguez-Rodríguez P, de Castro AGF, Sosa PA (2018) The restoration of the endangered Sambucus palmensis after 30 years of conservation actions in the Garajonay National Park: genetic assessment and niche modeling. PeerJ 6:e4985. https://doi.org/10.7717/peerj.4985
Rossetto M, McNally J, Henry RJ, Hunter J, Matthes M (2000) Conservation genetics of an endangered rainforest tree (Fontainea oraria—Euphorbiaceae) and implications for closely related species. Conserv Genet 1:217–229. https://doi.org/10.1023/A:1011549604106
Rossetto M, Bragg J, Kilian A, McPherson H, van der Merwe M, Wilson PD (2019) Restore and renew: a genomics-era framework for species provenance delimitation. Restor Ecol 27:538–548. https://doi.org/10.1111/rec.12898
Rossetto M, Wilson PD, Bragg J, Cohen J, Fahey M, Yap JYS, van der Merwe M (2020) Perceptions of similarity can mislead provenancing strategies—an example from five co-distributed acacia species. Diversity 12:306. https://doi.org/10.3390/d12080306
Rossetto M, Yap JYS, Lemmon J, Bain D, Bragg J, Hogbin P, Gallagher R, Rutherford S, Summerell B, Wilson TC (2021) A conservation genomics workflow to guide practical management actions. Global Ecol Conserv 26:e01492. https://doi.org/10.1016/j.gecco.2021.e01492
Rutherford S, van der Merwe M, Wilson PG, Kooyman RM, Rossetto M (2019) Managing the risk of genetic swamping of a rare and restricted tree. Conserv Genet 20:1113–1131. https://doi.org/10.1007/s10592-019-01201-4
Sansaloni C, Petroli C, Jaccoud D, Carling J, Detering F, Grattapaglia D, Kilian A (2011) Diversity arrays technology (DArT) and next-generation sequencing combined: genome-wide, high throughput, highly informative genotyping for molecular breeding of eucalyptus. BMC Proc 5:54. https://doi.org/10.1186/1753-6561-5-S7-P54
Schoen DJ, Brown AHD (1993) Conservation of allelic richness in wild crop relatives is aided by assessment of genetic markers. Proc Natl Acad Sci 90:10623–10627. https://doi.org/10.1073/pnas.90.22.10623
Schwartz M, Luikart G, Waples R (2007) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22:25–33. https://doi.org/10.1016/j.tree.2006.08.009
Shaffer HB, Toffelmier E, Corbett-Detig RB et al (2022) Landscape genomics to enable conservation actions: the California conservation genomics project. J Hered 113:577–588. https://doi.org/10.1093/jhered/esac020
Shapcott A, Lamont RW, O’Connor KM, James H, Conroy GC (2015) Population genetics of Philotheca sporadica (Rutaceae) to advise an offset translocation program. Conserv Genet 16:687–702. https://doi.org/10.1007/s10592-014-0693-x
Silcock JL, Simmons CL, Monks L, Dillon R, Reiter N, Jusaitis M, Vesk PA, Byrne M, Coates DJ (2019) Threatened plant translocation in Australia: a review. Biol Cons 236:211–222. https://doi.org/10.1016/j.biocon.2019.05.002
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Natl Acad Sci 101:15261–15264. https://doi.org/10.1073/pnas.0403809101
Supple MA, Shapiro B (2018) Conservation of biodiversity in the genomics era. Genome Biol 19:1–12. https://doi.org/10.1186/s13059-018-1520-3
Swofford DL, Sullivan J (2012) Phylogeny inference based on parsimony and other methods using PAUP. In: Lemey P, Salemi M, Vandamme A-M (eds) The phylogenetic handbook: a practical approach to phylogenetic analysis and hypothesis testing, 2nd edn. Cambridge University Press, Cambridge, pp 267–312
Theissinger K, Fernandes C, Formenti G, Bista I, Berg PR, Bleidorn C, Bombarely A, Crottini A, Gallo GR, Godoy JA, Jentoft S (2023) How genomics can help biodiversity conservation. Trends Genet. https://doi.org/10.1016/j.tig.2023.01.005
Turvey ST, Crees JL (2019) Extinction in the anthropocene. Curr Biol 29:982–986
Van Rossum F, Hardy OJ (2022) Guidelines for genetic monitoring of translocated plant populations. Conserv Biol 36:e13670. https://doi.org/10.1111/cobi.13670
Van Rossum F, Raspé O (2018) Contribution of genetics for implementing population translocation of the threatened Arnica montana. Conserv Genet 19:1185–1198. https://doi.org/10.1007/s10592-018-1087-2
Varshney RK, Roorkiwal M, Sun S et al (2021) A chickpea genetic variation map based on the sequencing of 3,366 genomes. Nature 599:622–627. https://doi.org/10.1038/s41586-021-04066-1
Willi Y, Kristensen TN, Sgrò CM, Weeks AR, Ørsted M, Hoffmann AA (2022) Conservation genetics as a management tool: the five best-supported paradigms to assist the management of threatened species. Proc Natl Acad Sci 119:e2105076119. https://doi.org/10.1073/pnas.2105076119
Wilson TC, Conn BJ, Henwood MJ (2012) Molecular phylogeny and systematics of Prostanthera (Lamiaceae). Aust Syst Bot 25:341–352. https://doi.org/10.1071/SB12006
Zheng X, Zheng MX (2013) Package ‘SNPRelate’. A package for parallel computing toolset for relatedness and principal component analysis of SNP data. Available online. http://github.com/zhengxwen/SNPRelate
Acknowledgements
We would like to acknowledge the Traditional Custodians of the land on which all plant species in this study are found, and pay respects to Elders past, present and emerging. This work was funded by the Saving Our Species program of the Department for Planning and Environment (DPE, New South Wales, Australia). We also acknowledge all the DPE threatened species officers and accompanying collectors that participated in the sampling and overall management of the species in this study, and the reviewers for their valuable suggestions.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by the New South Wales Government’s Saving Our Species conservation programme. The authors have no relevant financial or non-financial interests to disclose.
Author information
Authors and Affiliations
Contributions
MR conceived the study and led the write-up. MR, DB, SY obtained the funding supporting the research. JB, TW, SY contributed the analytical approaches and datasets. All authors participated in the writing and editing of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
Not applicable.
Additional information
Communicated by Thomas Abeli.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rossetto, M., Bragg, J., Brown, D. et al. Applying simple genomic workflows to optimise practical plant translocation outcomes. Plant Ecol 224, 803–816 (2023). https://doi.org/10.1007/s11258-023-01322-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11258-023-01322-4