Abstract
Species co-occurrences in local communities can arise independent or dependent on species’ niches. However, the role of niche-dependent processes has not been thoroughly deciphered when generalized to biogeographical scales, probably due to combined shortcomings of data and methodology. Here, we explored the influence of environmental filtering and limiting similarity, as well as biogeographical processes that relate to the assembly of species’ communities and co-occurrences. We modelled jointly the occurrences and co-occurrences of 1016 tropical tree species with abundance data from inventories of 574 localities in eastern South America. We estimated species co-occurrences as raw and residual associations with models that excluded and included the environmental effects on the species’ co-occurrences, respectively. Raw associations indicate co-occurrence of species, whereas residual associations indicate co-occurrence of species after accounting for shared responses to environment. Generally, the influence of environmental filtering exceeded that of limiting similarity in shaping species’ co-occurrences. The number of raw associations was generally higher than that of the residual associations due to the shared responses of tree species to the environmental covariates. Contrary to what was expected from assuming limiting similarity, phylogenetic relatedness or functional similarity did not limit tree co-occurrences. The proportions of positive and negative residual associations varied greatly across the study area, and we found a significant tendency of some biogeographical regions having higher proportions of negative associations between them, suggesting that large-scale biogeographical processes limit the establishment of trees and consequently their co-occurrences.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The immense diversity of tropical tree communities and its drivers have intrigued scientists for decades. Research on the topic has focused on processes allowing species to occur together despite the limited resources available for growth and reproduction (e.g., Chesson 2000; Diamond 1975; Hardin 1960). Studying tropical tree co-occurrences can reveal the scale-dependent effects of the ecological and biogeographical processes underlying the observed patterns. In general, community assembly processes govern how species in a regional pool are distributed into local communities, and thereby determine the co-occurrences of species within the local communities (MacArthur and Levins 1967; van der Valk 1981). Here, we consider two species to co-occur biologically meaningfully and statistically significantly if they occur in the same forest study plot more frequently or in larger abundances than assumed by random (Ovaskainen et al. 2017).
At large spatial scales, species’ co-occurrences are a function of processes that can give rise to regional and biogeographical differences in co-occurrence patterns. Chance biogeographical events, such as presence of dispersal vector, distance to new environments (Lortie et al. 2004), time since last glacial period (Adams and Woodward 1989) or continental drift-induced distributions of major taxonomic lineages, can influence which species are capable of reaching the focal environment. Indeed, increased regional species richness can result only from dispersal of species into the region or from in situ speciation, processes that are best identified using historical biogeography (Wiens and Donoghue 2004). Furthermore, priority effects, i.e., randomly determined order of species’ arrival to the local community, may affect the final composition of the community (Fukami et al. 2005). The majority of studies on species co-occurrence patterns are conducted at the scale of single forest patches and not generalized to larger spatial extents (e.g., McFadden et al. 2019; Seidler and Plotkin 2006; Wiegand et al. 2007, but see Kunstler et al. 2012; Zambrano et al. 2017). Therefore, linking local and regional community dynamics as well as ecological and biogeographical processes in generating co-occurrences is essential to understand species’ co-occurrences in large spatial context.
At local spatial scales, the presence and abundance of a species in a community are a function of the properties of its niche. The niche-based processes can promote either convergence (environmental filtering) or divergence (limiting similarity) in traits (MacArthur and Levins 1967; van der Valk 1981). Environmental filtering is a niche-based process that excludes species from the community if their niches are not suited to the local environmental conditions (van der Valk 1981; Keddy 1992). Many abiotic factors may constrain species co-occurrences through environmental filtering, including climatic and soil properties (as reviewed by Kraft et al. 2015). Limiting similarity is a niche-based process that prevents species from co-occurring in a community if their niches are too similar, due to competitive exclusion (MacArthur and Levins 1967). Theoretically, species with the same set of life-history traits are expected to compete and not to co-occur in space and time (Kraft et al. 2008; Wilson and Stubbs 2012). However, competition may also sometimes prevent coexistence of dissimilar taxa (Mayfield and Levine 2010). Indeed, niche-based processes have gained criticism for being difficult to differentiate because they may produce similar co-occurrence patterns (Cadotte and Tucker 2017). Despite the clear limitations, niche-based processes and observational data have distinct value for inferring the role of the environment and species characteristics in community structure at large spatial scales as long as the results are interpreted with care.
In this paper, we study the processes underlying species’ co-occurrences at different spatial scales. Using comprehensive data on tropical tree abundances across a large spatial context, we investigate how environmental filtering and limiting similarity may affect the co-occurrence patterns in species-rich tree communities of tropical and subtropical regions in eastern South America. To understand the processes behind co-occurrences, we ask three specific questions. (1) Does the abiotic environment constrain the co-occurrences of tropical tree species? Following the preceding literature on patterns within single forest patches (e.g., Silva and Batalha 2009), we expected abiotic environment (including climate, disturbance, soil, and topography) to be important in constraining species occurrences and co-occurrences at large spatial scales. (2) Does phylogenetic relatedness or functional similarity limit the co-occurrences of tropical tree species? According to the limiting similarity hypothesis, we hypothesized that functionally similar or closely related species occur together less frequently and in lower abundances than expected due to niche overlap. We also expected that the traits of species with the strongest positive or negative co-occurrences with other species differ from the traits of species with weaker co-occurrences. (3) Do the co-occurrences of tropical tree species vary spatially across biogeographical regions? Since major biogeographical regions have substantial differences in their vegetation structure and composition (Olson et al. 2001) and in their biogeographical history, such as time since last glacial period (Adams and Woodward 1989; Segovia and Armesto 2015), we expected to find more negative co-occurrences among regions than within.
Methods
Data
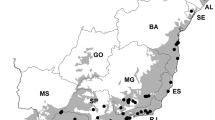
The studied tree communities are located in various tropical and subtropical regions in eastern South America, including the Atlantic Forest, Caatinga, Cerrado, Pampa, and Pantanal ecoregions (Fig. 1; Olson et al. 2001). The biogeographical history of South America produced areas of high speciation as well as high extinction rates due to continental drift, dispersal barriers, and new environmental conditions by the Andean uplift (Segovia and Armesto 2015). Eastern South America is characterized by a North–South gradient in precipitation seasonality, mean annual precipitation (mm), and minimum temperature (°C). These climatic gradients coupled with the variation in geomorphologic and edaphic conditions result in a wide spectrum of woodland types, ranging from tall rainforests to open canopy savannas. The study region includes forests that grow in a wide range of altitudes (0–2300 m.a.s.l.) and have varying proportions of deciduous trees and different soil properties. The major land cover types include shrubland, cropland, herbaceous vegetation, and forests (Buchhorn et al. 2019). The intensity of human influence increases towards the coast (Wildlife Conservation Society and Center for International Earth Science Information Network 2005), where the major urban areas are located.
Map of the hierarchically structured sampling design in eastern South America. Included levels are ecoregion (N = 10 (color); simplified based on Olson et al. 2001) and sampling site (N = 574; black circle). Distribution of sampling sites among ecoregions is indicated next to the legend
Species occurrences
We retrieved abundance data of 1016 tree species from 574 community surveys (totaling 961,184 individuals) from the Neotropical Tree Community database (TreeCo; http://labtrop.ib.usp.br/doku.php?id=projetos:treeco:start) using the methods described in Lima et al. (2015) and de Lima et al. (2020). For this specific study, we selected the surveys including trees from the dominant/adult stratum of the vegetation, which were defined to include trees with diameter at breast height (DBH) ≥ 5 cm for closed canopy forests and DBH ≥ 3 cm or DGH (diameter at ground height) ≥ 5 cm for open canopy forests and savannas. We included only those surveys that met the following criteria: a minimum sampling effort of 0.4 ha, data published after 2000 and with a minimum 90 percent of trees identified to species level. We did not consider planted or early secondary forests (< 25 years since land abandonment and forest regrowth). Furthermore, we selected only those surveys for which the geographical coordinates stored in TreeCo were validated at the forest fragment level, i.e., those for which the coordinates fell within the focal forest fragment where the survey was conducted. Within these surveys, we further selected species for which functional trait data were available, and which had a minimum of six occurrence records. The latter selection was done because our focus was on species’ co-occurrences that cannot be estimated reliably for species with very few occurrences (Ovaskainen and Abrego 2020). Of the initial 2932 species, 1616 had missing trait values and 1513 had fewer than six occurrence records, resulting in 1016 species (~ 35% of all species, ~ 80% of all individuals) included in the final analyses. Therefore, the species included in the analyses mainly cover well-studied species and genera that have available information of their traits. We defined trees as those plants with free-standing stems that can grow at least 4 m tall, including trees, treelets, palms, tree ferns, and cacti (for the list of included species, see Appendix S1). All species names were checked for typographical errors, synonyms, and orthographical variants following the Brazilian Flora 2020 nomenclature (Ranzato Filardi et al. 2018).
Environmental covariates and spatial structure
To study the possible abiotic effects on species co-occurrences patterns, we obtained climate, topography, soil and landscape covariates for each survey based on the spatial coordinates of the survey (for detailed definitions, spatial resolutions, and variation among study sites, see Appendix S2: Table S1, de Lima et al. 2020). Following preliminary analyses, we selected a set of uncorrelated variables to avoid collinearity in the model fitting and excluded those variables that were highly correlated (Pearson’r >|0.5|) with other variables (e.g., altitude). Climate covariates consisted of mean annual precipitation (mm) (Alvares et al. 2015; Fick and Hijmans 2017), mean annual temperature (°C) (Alvares et al. 2013; Fick and Hijmans 2017), and bioclimatic stress measured as a function of temperature seasonality, precipitation seasonality, and climatic water deficit (the ‘E’ parameter in Chave et al. 2014). As topography covariates, we included slope declivity (0°–90°) and aspect (northness and eastness, calculated, respectively, as cosine and sine of aspect), built based on 2000 NASA Shuttle Radar Topography Mission using GDAL-QGIS software (version 3.4.4). As a soil property measure, we used the soil quality information from the Harmonized World Soil database (version 1.2) to obtain soil quality index calculated semi-quantitatively as the sum of nutrient availability, nutrient retention capacity, rooting conditions, oxygen availability to roots, and excess salts (Fischer et al. 2008). Soil information that are adequate for studying crop plants (soil workability and toxicity) were not considered for calculating the soil quality index. Since each of the five included variables varied from 1 (no or slight limitations) to 4 (very severe limitations), the soil quality index varied from 4 to 16.
To account for the effects of forest patch size and human-induced disturbances, we obtained the area of the forest fragment surveyed (ha) and the human influence index (Sanderson et al. 2002), which varied from 0 (low human influence: wild areas) to 65 (high human influence: urban areas). Area of the fragment was obtained from the information in the original publication and cross-checked using the SOS Mata Atlântica/INPE Atlantic Forest fragments mapping (Fundação SOS Mata Atlântica 2014). We did not include landscape forest cover due to its strong correlation with forest fragment size that better corresponds to the local patch quality. Finally, we included the sampling effort (ha) and the sampling method (point-centered quadrant, plot) as reported in the original publication to account for potential sampling effects. We note that the spatial and temporal resolution of the included covariates varies, and thus they may be coarse in some cases, which may dilute their modelled effects on species’ occurrences and co-occurrences. However, these variations in the spatial and temporal resolutions are likely to contribute to increased uncertainty, rather than bias, in the model estimates.
We compiled species occurrence data hierarchically at ecoregion and sampling site scales (Fig. 1). At the larger scale, we included ecoregions without spatial coordinates. Ecoregions were obtained and simplified from the Nature Conservancy (TNC) definitions (ecoregion scale, N = 10; Olson et al. 2001). Although the ecoregions, such as Cerrado and Caatinga, are distinguished from each other by biotic and abiotic differences, the borders between them are not discrete, and transitional zones of varying extents generally exist between the regions. At the smaller scale, we included the local hierarchical level of the sampling site with its spatial coordinates (site scale, N = 574).
Species characteristics
To study the effect of shared evolutionary history on species co-occurrence patterns, we built the phylogenetic tree based on the stored megatree R20120829 from Phylomatic (version 3; http://phylodiversity.net/phylomatic). The tree was calibrated using ‘bladj’ algorithm in Phylocom software (Webb et al. 2008), which is based on node ages suggested by Bell et al. (2010) and Magallón et al. (2015). We eliminated polytomies by generating random dichotomies with length 0.001 between sister species. To solve polytomies we used the ‘ape’ package in R software (version 3.5.0; Paradis et al. 2004). Finally, we constructed a matrix of evolutionary distances in million years across all species pairs. We note that the phylogeny was compiled based on fossil records due to the limited availability of phylogenies built from DNA data. Nonetheless, the phylogeny suffices for detecting patterns of phylogenetically structured co-occurrences among a large number of species pairs across ecoregions.
To assess the effect of functional similarity on species’ co-occurrences, we obtained from TreeCo database those plant traits that reflect the major axes of variation in ecological strategies and are thus relevant for assessing niche-based processes (Díaz et al. 2015; Lopez et al. 2016; Martins et al. 2018). These included seed length (cm), wood density (g/cm3), maximum growth height (m), leaf area (cm2), leaf type (compound, simple), dispersal syndrome (autochoric, anemochoric, barochoric, hydrochoric, zoochoric), and successional group (pioneer, initial/late secondary, climax). In addition, we included the geographic distribution (local/regional endemic, central/southern/northern/western South America, Neotropical, Pantropical, exotic). The details on obtaining species’ characteristics can be found in de Lima et al. (2020). Based on the assumption that closely related species tend to have similar trait values, we completed the trait matrix with genus level averages in cases of species missing values of seed length, wood density, and dispersal syndrome. We calculated a pairwise trait distance matrix using Gower distances in 'FD' package (Laliberté and Legendre 2010) in R software (version 3.5.0; R Core Team 2019), thus allowing inclusion of categorical traits.
Statistical analyses
Joint species distribution modelling
We synthesized data on species occurrences and abundances, and environmental and spatial variables using the Hierarchical Modelling of Species Communities framework (HMSC; Ovaskainen et al. 2017; Ovaskainen and Abrego 2020). HMSC is a class of joint species distribution models (Warton et al. 2015), which allows simultaneous modelling of the occurrences/abundances of all species as a function of their responses to the environmental covariates. Of particular relevance for addressing the main objective of the present study, HMSC employs a latent variable approach that allows estimating pairwise residual species’ associations (i.e., statistically supported species-to-species associations that remain after accounting for the effects of the environmental covariates from species-rich community data) (Ovaskainen et al. 2016b).
To account for the zero-inflated nature of the data, the models were fitted by following a hurdle modelling approach in which we modelled separately the presence–absences and the abundances conditional on presence (Cragg 1971; Ovaskainen and Abrego 2020). This approach allows disentangling the ecological processes that explain the variation in species’ presence–absences from those explaining their abundances. We applied probit regression for the presence–absence data and log-normal regression for the abundance data conditional on presence. We fitted in total four HMSC models, corresponding to two alternative sets of the hurdle-type models. In the first set of hurdle models, we accounted for the spatial structure of the data through (latent variable) random effects at the sampling site and ecoregion levels, but we did not include environmental covariates. The second set of hurdle models was otherwise identical to the first set, but included the environmental covariates explained in the previous section (describing the climate, topography, soil, landscape, and sampling design). These two sets of models allowed us to estimate the raw and residual species-to-species associations, respectively. The raw associations represent the overall pairwise associations among the species disregarding which factors drive the co-occurrences. For the residual associations, the species’ shared responses to the environmental covariates included in the models are accounted for. As explained below in more detail, for all four models we estimated the species-to-species associations at the scales of sampling sites and ecoregions. The models were fitted within the Bayesian inference framework using the MATLAB implementation of HMSC and the default prior distributions (the MATLAB HMSC software and manual are found at https://www.helsinki.fi/en/researchgroups/statistical-ecology/hmsc). We calculated the proportions of positive and negative associations among all species pairs with at least 95 percent posterior probability across the posterior samples for each of the four models.
Modelling species-rich communities is generally challenging as computation times increase exponentially with an increasing number of species. HMSC allows circumventing this problem with a latent variable approach that can be viewed as a model-based ordination (Warton et al. 2015; Ovaskainen et al. 2016a, 2017). The tree species were modelled with a set of shared latent factors that have species-specific loadings (Ovaskainen et al. 2016a, b). In HMSC, the number of latent factors is adapted in the model fitting procedure so that a sufficient number of latent factors are ensured to capture the ecologically relevant variation and to avoid modelling noise rather than actual signal (Ovaskainen et al. 2016a, b). We included latent factors at the ecoregion (including ecoregion identity as a random effect) and sampling site (including sampling site coordinates as a spatially structured random effect) levels to account for the spatial autocorrelation in the species occurrence/abundance data. Thus, the ecoregion and sampling site level latent factor structure correspond to the random effects part of the models. With the latent variable approach, the species-to-species variance–covariance matrix (hereafter referred to as the association matrix) can be represented through latent factors and their loadings (Ovaskainen et al. 2017). The factor loadings indicate patterns where a pair of species co-occur less or more frequently or in higher or lower abundances than expected given the other parameters in the fitted models. That is, with loadings of the same sign, the pair increase similarly in occurrence probability or abundance, whereas with loadings of opposite signs, one species declines and the other increases (Ovaskainen et al. 2017). However, we note that this is a correlative approach and the results strongly depend on whether the relevant covariates have been included in the models. Thus, a proper interpretation of the results requires high-quality data of the relevant environmental covariates. For more details and examples on the latent variable approach, see Chapter 7 in Ovaskainen and Abrego (2020).
We evaluated the explanatory and predictive powers of the models by calculating Tjur’s R2 (Tjur 2009) in the case of presence–absence data, and by calculating the correlation between the observed and predicted abundances in the case of the abundance conditional on presence data. To compute explanatory power, we made model predictions based on models fitted to all data. To compute predictive power, we applied the following fourfold cross-validation approach. We divided the study sites randomly into four sets, fitted the model using three of the four sets as training data, and predicted the validation data on the remaining fourth set of sites. We repeated this analysis four times, thus generating an independent prediction for each site.
Estimating effects of environmental filtering on species co-occurrences (Question 1)
To answer the first research question on abiotic effects on co-occurrences, we evaluated the relative role of environmental filtering by comparing the proportions of positive and negative associations between the raw and residual association matrices. For all models, we partitioned the explained variance in species occurrences and co-occurrences among the included environmental predictors and random effects.
Estimating effects of phylogenetic relatedness and functional similarity on species co-occurrences (Question 2)
To answer the second research question on the biotic effects on co-occurrences, we calculated the correlations between the pairwise raw and residual species-to-species associations the pairwise phylogenetic and trait distances. For this, we applied a Mantel test with 1000 permutations. As opposed to the phylogenetic niche conservatism hypothesis (Harvey and Pagel 1991), there was a statistically significant but negligible correlation between pairwise phylogenetic and trait distances (Mantel r = 0.04, p = 0.003). When testing the correlation for phylogenetic distances and distances of each continuous trait separately, we did not find significant correlations. Thus, we treated phylogenetic relatedness as an independent factor in the analyses, rather than as a proxy for species’ functional space. According to the limiting similarity hypothesis, we expected to observe a negative correlation between species’ co-occurrences and similarity. However, we acknowledge that competitive interactions may also involve competitive hierarchies wherein certain traits are competitively advantageous, leading to trait convergence (Mayfield and Levine 2010).
Finally, we compared the trait distributions of species with the strongest positive (N = 20) and negative (N = 20) residual associations to assess whether they represented distinctive trait combinations.
Assessing spatial distribution of species co-occurrences (Question 3)
To answer the third research question on spatial variation in co-occurrences, we calculated for each of the local tree communities the percentages of positive and negative residual associations as sum of all statistically supported associations of species pairs. Based on the spatial distribution of the co-occurrences, we estimated the potential presence of biogeographical processes. That is, we assumed that the variation in co-occurrence patterns among ecoregions to reflect biogeographical processes. However, we note that also other processes, such as processes related to species’ interactions, may also lead to large-scale spatial variation in co-occurrences.
We plotted the percentage of associations across eastern South America and applied a parametric analysis of variance test (ANOVA) to study differences in mean percentages among ecoregions. We applied a non-parametric analysis of variance (Kruskal–Wallis) when the assumptions for the parametric analysis of variance test were not met (Appendix S2, Fig. S1). Furthermore, we assessed whether the transitional zones between ecoregions exhibited distinct percentages of positive and negative associations. We defined the transitional zones as those areas within a close proximity to established limits of ecoregions, where the communities are likely to exhibit characteristics of both ecoregions (McDonald et al. 2005; Smith et al. 2018). However, the transitional zones likely vary in extent among the ecoregions and should be considered as suggestive, rather than definitive.
Results
Model fit and estimated co-occurrences
The models fitted to the presence–absence data without and with environmental covariates explained 18.9 percent and 25.1 percent of the variation at the sampling site level, respectively. The predictive power based on cross-validation was 12.3 percent for the model without environmental covariates and 12.6 percent for the model with environmental covariates. The models fitted to the abundance data without and with environmental covariates explained 60.6 percent and 72.0 percent of the variation in species’ abundances at the sampling site level, respectively. The predictive power based on cross-validation was 16.3 percent for the model without environmental covariates and 25.8 percent for the model with environmental covariates. Note that different types of R2 measures were used between the models fitted to the presence–absence and abundance data, so that these numbers are not comparable as such.
Based on the loadings obtained with the latent factor approach, we estimated more positive than negative statistically supported associations. However, the estimated proportions of positive and negative associations differed between the studied spatial scales and between the models fitted to presence–absence and abundance data, as well as between models fitted without and with environmental covariates (Table 1). The observed associations were largely different at site and ecoregion scales, likely encompassing local assembly processes and biogeographical processes, respectively. Overall, we estimated more associations based on the models fitted to the presence–absence than to the abundance data. Furthermore, we estimated more associations at the sampling site level than at the ecoregion level.
Effects of environmental filtering on species co-occurrences
Both models fitted to the presence–absence and to the abundance data explained species’ co-occurrences better when the environmental factors were accounted for. However, for the presence–absence data, the cross-validated predictive powers did not differ between models including and excluding environmental covariates. For the abundance data, the models including environmental covariates estimated fewer positive and negative associations than the models without environmental covariates (Table 1).
In the presence-absence model, the included environmental covariates explained 32 percent of the explained variation in species’ occurrences, whereas the site and ecoregion random effects explained 66 percent (Table 2). In the abundance conditional on presence model, the included environmental covariates explained 41 percent of the explained variation in species’ abundances, while the site and ecoregion random effects explained 42 percent (Table 2). In both models, the included climatic factors (mean annual precipitation, mean annual temperature, and bioclimatic stress) were the most important environmental covariates explaining on average 25 percent of the total explained variation (Table 2). The effect of forest fragment area and human influence was larger on tree abundances than presence–absences (Table 2).
Effects of phylogenetic relatedness and functional similarity on species co-occurrences
Mantel tests showed no statistically significant correlations between the raw association and phylogenetic and trait distance matrices, except for the presence–absence data and phylogenetic distances (test results reported within Fig. 2). However, the correlative relationship between phylogenetic distances and the raw associations was rather weak. The correlations between the residual association and phylogenetic and trait distance matrices were similar to those computed for the raw associations (Appendix S2, Table S2).
Relationships of pairwise raw association strengths and phylogenetic and trait distances according to the models fitted to the presence–absence data (a, b) and abundance data (c, d). a, b Represent the relationships of raw association strength–phylogenetic distance and raw association strength–trait distance according to the model fitted to the presence–absence data, respectively. c, d Represent the relationships of raw association strength–phylogenetic distance and raw association strength–trait distance, respectively. Each colored hexagon represents the number of points that fall within it, while each point represents the value of an estimated pairwise association (the darker the shade of grey, the higher the number of points within the hexagon). Mantel test results (correlation coefficient (r) and significance (p)-values) based on 1000 permutations are shown for each matrix pair correlation
According to the model fitted to the abundance data, the species with the strongest positive associations was Zanthoxylum rhoifolium, whereas the species with the strongest negative associations was Guettarda viburnoides (see Appendix S2, Table S4 for the full lists of the species with the strongest associations). The trait spaces of the species with the strongest positive (20 species) and negative associations (20 species) did not differ significantly from each other (Appendix S2, Table S5).
Spatial configuration of species co-occurrences
According to the model fitted to the abundance data, we found the highest proportions of residual positive associations in Alto Parana and Uruguayan Savanna ecoregions, and the differences among the ecoregions in general were statistically significant (parametric ANOVA: F = 54.6, df = 572, p < 0.01; Fig. 3; Appendix S2, Fig. S2). The proportions of residual negative associations were highest in Cerrado ecoregion and its transitional zones with other ecoregions, and the differences were statistically significant among the ecoregions (non-parametric Kruskal–Wallis: χ2 = 249.44, df = 9, p < 0.001; Fig. 3; Appendix S2, Table S3).
Spatial distribution of proportions of (a) residual negative and (b) residual positive associations over the species pairs present across the sampling sites and the variation of (c) residual negative and (d) residual positive association proportions in each ecoregion, that are delimited with grey lines in (a, b) (see Fig. 1 for the ecoregion names). The transitional zones between the ecoregions are not indicated as they may vary in extent across the study area. Results are based on the model fitted to the abundance data. Note the different y-axis scales in (c, d)
Discussion
Our study using a comprehensive dataset of tropical trees shows that large-scale tree co-occurrence patterns are determined by environmental filtering and biogeographical processes more than by limiting similarity. Environmental filtering effects explained more variation for species’ abundance than presence–absence data, indicating that environmental variables explained better whether tree species were abundant than where they occurred. Moreover, including covariates in the models generally improved their explanatory powers, and the covariates explained many of the tree co-occurrences. However, at large scales, we found a large spatial variation in species' co-occurrence patterns. Hence, our findings suggest that the niche-based processes govern the local co-occurrence patterns whereas biogeographic processes govern the large-scale co-occurrence patterns.
Of all environmental covariates, the variation in species occurrences was best explained by the climatic variables, including mean temperature and precipitation, as well as climate seasonality. In addition, we found that the effect of anthropogenic disturbances (here, forest fragment area and human influence) was larger on tree abundances than presence–absences, which may indicate declining population trends for some species and increasing population trends for others under intensifying anthropogenic pressures. This suggests that climate change may alter tree species’ spatial distributions (similarly to Miles et al. 2004), while anthropogenic disturbances may alter species’ relative abundance distributions. However, due to the varying spatial and temporal resolution of the environmental data, the environmental variables, such as soil quality, may not be able to capture the full extent of the environmental effects on species occurrences. Moreover, by excluding the non-adult individuals and the rarest species from the analyses (i.e., including ~ 35% of the original number of observed species and ~ 80% of the original number of observed individuals), we may have overlooked some of the environmental filtering and limiting similarity effects on the rarer species. Therefore, it is possible that environmental filtering is an even more important driver of occurrence and co-occurrence patterns. In addition to the environmental variables, the variation in both species’ presence–absences and abundances was largely explained by the spatially structured random effects, suggesting that species’ occurrences were spatially structured along gradients of unmeasured covariates, such as land use intensity or time since last glacial period. Moreover, a significant proportion of the variation in species’ abundances was explained by the sampling effort, as the number of observed individuals logically tends to increase with increasing sampling effort. To disentangle the importance of actual environmental variables, it is important to control for such sampling design-dependent effects.
According to the limiting similarity hypothesis, co-occurrences among phylogenetically closely related and functionally similar species should be predominantly negative. Contrary to previous research (Kraft et al. 2008; Wilson and Stubbs 2012, but see Silva and Batalha 2009), we did not observe any signs of limiting similarity at the spatial extent of our study. One explanation for this might be that limiting similarity is only important at very fine spatial scales, whereas at larger spatial scales other processes cancel its effects. For instance, when considering species’ presence–absences, competitive exclusion can take an extremely long time and the importance of limiting similarity in that may be overridden by speciation (Hubbell and Foster 1986), leading to random patterns of species co-occurrences. At the spatial scale of our study, outcomes of limiting similarity may be masked because we considered species occurrences without information of the spatial configuration of individual trees within the sites. Therefore, the modelled associations may not reflect the fine scale avoidance of similar species as they may still co-occur within the same sampling site. Future research could use our approach to model residual co-occurrences with individual-based data below plot scales, thus allowing the assessment of limiting similarity effects at much finer spatial scales.
Tree communities with the highest proportions of negative associations were located in the transitional zones between the major biogeographical regions (e.g., savanna-seasonal forest-transition), suggesting a dispersal and/or establishment barrier between regions. Because we modelled species’ associations based on abundance data conditional on presence, species’ co-occurrence within sampling sites are implicit. Thus, we expect negative associations from the model fitted to the abundance data to reflect establishment rather than dispersal barriers. Moreover, 75% of the studied species are animal-dispersed, a dispersal syndrome known to be efficient (Myers et al. 2004), making dispersal limitation the less plausible mechanism. Rare long distance dispersal events may be key to the colonization of new ecoregions (Clark et al. 1999, 2005). Thus, tree occurrences are mainly driven by establishment and growth, which are affected by many ecological factors, such as seed predation and light conditions (Janzen 1970; Rüger et al. 2011). Transitional zones between ecoregions are highly variable and may induce small-scale spatial variation in species’ co-occurrences. For example, the transitional zone between Cerrado and Caatinga is likely to stem from their difference in the length of the dry season, whereas the transitional zones between Bahia, Serra do Mar, and Araucaria are likely founded on temperature differences (Liebmann et al. 2007; Alvares et al. 2013). Indeed, many species occur at their range limits in the transitional zones, leading to co-occurrence patterns consistent with environmental filtering (Sommer et al. 2014). An alternative explanation for the spatial variation in residual associations along transitional zones is that they represent ecological interactions (Sommer et al. 2014). We note that using co-occurrences as direct proxies for pairwise interactions is problematic (Dormann et al. 2018; Freilich et al. 2018). However, indirect species interactions, such as apparent competition, are few in the literature and research tends to focus on the observed networks of direct interactions. As a result, significant associations are often disregarded as false positives or negatives in co-occurrence analyses (e.g., Freilich et al. 2018). Thus, the estimated residual associations pose interesting hypotheses about direct and indirect ecological interactions to be tested in the future research. Finally, fitting separate models to presence–absence and abundance data yielded additional evidence for the existence of biogeographical scale mechanisms that lead to spatial variation in the association patterns: there were more negative associations for the presence–absence data than for the abundance data. This suggests that it is more common for species to affect each other’s occurrences than abundances at the scale of our study.
Here, we studied the influence of environmental filtering and limiting similarity on species’ co-occurrences at a site level. However, one cannot fully separate different niche-based processes based solely on co-occurrences patterns, because functional niche differences are influenced by both environmental and competitive factors (Kraft et al. 2015). Detection of limiting similarity may be particularly challenging if the traits that drive local coexistence are also the same traits that drive competitive exclusion, or if only certain combinations of traits are reflective of competitive effects (Kraft et al. 2015). While the environment can filter functionally similar species into the local species pool (Bazzaz 1991; Kraft et al. 2015), competition may allow the local co-occurrence of functionally similar species (Chesson 2000; Mayfield and Levine 2010). For example, particular plant traits are important for adaptation to the local environmental conditions independent of the species (Díaz et al. 2015). Thus, species in distant lineages may express high functional similarity (Swenson and Enquist 2009). Variation in these niche-based processes may lead to functional differences among the local communities at larger spatial scale if the environment is filtering groups of species that are functionally similar (Hérault 2007). Thereby, environmental filtering through climate and habitat characteristics may select for a set of discrete common characteristics that differ between biogeographical regions (Echeverría-Londoño et al. 2018; Li et al. 2018).
Understanding how the abiotic environment drives tree species’ occurrences and co-occurrences has both conservational and methodological applications. Firstly, knowing which species tend to co-occur has potential of informing holistic conservation and restoration efforts so that tightly linked species can be protected and/or reintroduced together using similar environmental assumptions. Secondly, shifts in tree occurrences due to environmental factors need to be accounted for in conservation prioritization as future distributions of species may not match the current ones (Miles et al. 2004). Thirdly, presence–absence data alone may not suffice for inferring the effects of environmental change on species communities as the negative population trends may be masked until (local) extinctions of species unless abundance data are obtained. Finally, when assessing co-occurrence patterns at large spatial scales, including environmental covariates in the model is essential. Otherwise, estimated raw co-occurrences will largely represent species’ shared responses to the abiotic environment rather than ecologically meaningful co-occurrences.
In this paper, we found that tree species’ co-occurrences are mostly determined by environmental and sampling factors, more than by species’ phylogenetic relatedness or functional similarity. The co-occurrence patterns also greatly varied across the study region, which indicates presence of underlying spatially structured, biogeographical processes. These results highlight the need for studies at large spatial scales, as they can provide additional information on the hierarchical processes that shape species' co-occurrences.
Data availability
The utilized data are stored in Neotropical Tree Communities (TreeCo) database and are available upon request through the database website.
Code availability
The code and manual for fitting the HMSC models are available at https://www.helsinki.fi/en/researchgroups/statistical-ecology/hmsc.
References
Adams JM, Woodward FI (1989) Patterns in tree species richness as a test of the glacial extinction hypothesis. Nature 339:699–701. https://doi.org/10.1038/339699a0
Alvares CA, Stape JL, Sentelhas PC, de Moraes Gonçalves JL (2013) Modeling monthly mean air temperature for Brazil. Theor Appl Climatol 113:407–427. https://doi.org/10.1007/s00704-012-0796-6
Alvares CA, de Mattos EM, Sentelhas PC, Miranda AC, Stape JL (2015) Modeling temporal and spatial variability of leaf wetness duration in Brazil. Theor Appl Climatol 120:455–467. https://doi.org/10.1007/s00704-014-1182-3
Bazzaz FA (1991) Habitat selection in plants. Am Nat 137:116–130. https://doi.org/10.1086/285142
Bell CD, Soltis DE, Soltis PS (2010) The age and diversification of the angiosperms re-revisited. Am J Bot 97:1296–1303. https://doi.org/10.3732/ajb.0900346
Buchhorn M, Smets B, Bertels L, Lesiv M, Tsendbazar N-E, Herold M, Fritz S (2019) Copernicus global land service: land cover 100m: epoch 2015: globe. Dataset of the global component of the Copernicus Land Monitoring Service. https://land.copernicus.eu/global/documents/scientific
Cadotte MW, Tucker CM (2017) Should environmental filtering be abandoned? Trends Ecol Evol 32:429–437. https://doi.org/10.1016/j.tree.2017.03.004
Chave J, Réjou-Méchain M, Búrquez A, Chidumayo E, Colgan MS, Delitti WBC, Duque A, Eid T, Fearnside PM, Goodman RC, Henry M, Martínez-Yrízar A, Mugasha WA, Muller-Landau HC, Mencuccini M, Nelson BW, Ngomanda A, Nogueira EM, Ortiz-Malavassi E, Pélissier R, Ploton P, Ryan CM, Saldarriaga JG, Vieilledent G (2014) Improved allometric models to estimate the aboveground biomass of tropical trees. Glob Change Biol 20:3177–3190. https://doi.org/10.1111/gcb.12629
Chesson P (2000) Mechanisms of maintenance of species diversity. Annu Rev Ecol Syst 31:343–366. https://doi.org/10.1146/annurev.ecolsys.31.1.343
Clark JS, Silman R, Macklin E, HilleRisLambers J (1999) Seed dispersal near and far: patterns across temperate and tropical forests. Ecology 80:1475–1494. https://doi.org/10.1890/0012-9658(1999)080[1475:SDNAFP]2.0.CO;2
Clark CJ, Poulsen JR, Bolker BM, Connor EF, Parker VT (2005) Comparative seed shadows of bird-, monkey-, and wind-dispersed trees. Ecology 86:2684–2694. https://doi.org/10.1890/04-1325
Cragg JG (1971) Some statistical models for limited dependent variables with application to the demand for durable goods. Econometrica 39:829. https://doi.org/10.2307/1909582
de Lima RAF, Oliveira AA, Pitta GR, de Gasper AL, Vibrans AC, Chave J, ter Steege H, Prado PI (2020) The erosion of biodiversity and biomass in the Atlantic Forest biodiversity hotspot. Nat Commun 11:6347. https://doi.org/10.1038/s41467-020-20217-w
Diamond JM (1975) Assembly of species communities. In: Cody ML, Diamond JM (eds) Ecology and evolution of communities. Harvard University Press, Cambridge, pp 342–444
Díaz S, Kattge J, Cornelissen JHC, Wright IJ, Lavorel S, Dray S, Reu B, Kleyer M, Wirth C, Colin Prentice I, Garnier E, Bönisch G, Westoby M, Poorter H, Reich PB, Moles AT, Dickie J, Gillison AN, Zanne AE, Chave J, Joseph Wright S, Sheremetev SN, Jactel H, Baraloto C, Cerabolini B, Pierce S, Shipley B, Kirkup D, Casanoves F, Joswig JS, Günther A, Falczuk V, Rüger N, Mahecha MD, Gorné LD (2015) The global spectrum of plant form and function. Nature 529:167–171. https://doi.org/10.1038/nature16489
Dormann CF, Bobrowski M, Dehling DM, Harris DJ, Hartig F, Lischke H, Moretti MD, Pagel J, Pinkert S, Schleuning M, Schmidt SI, Sheppard CS, Steinbauer MJ, Zeuss D, Kraan C (2018) Biotic interactions in species distribution modelling: ten questions to guide interpretation and avoid false conclusions. Glob Ecol Biogeogr 27:1004–1016. https://doi.org/10.1002/adsc.201
Echeverría-Londoño S, Enquist BJ, Neves DM, Violle C, Boyle B, Kraft NJB, Maitner BS, Mcgill B, Peet RK, Sandel B, Smith SA, Svenning J-C, Wiser SK (2018) Plant functional diversity and the biogeography of biomes in North and South America. Front Ecol Environ 6:219. https://doi.org/10.3389/fevo.2018.00219
Fick SE, Hijmans RJ (2017) WorldClim 2: New 1-km spatial resolution climate surfaces for global land areas. Int J Climatol 37:4302–4315. https://doi.org/10.1002/joc.5086
Fischer G, Nachtergaele F, Prieler S, van Velthuizen HT, Verelst L, Wiberg D (2008) Global agro-ecological zones assessment for agriculture (GAEZ 2008). IIASA, FAO, Laxenburg, Rome
Freilich MA, Wieters E, Broitman BR, Marquet PA, Navarrete SA (2018) Species co-occurrence networks: can they reveal trophic and non-trophic interactions in ecological communities? Ecology 99:690–699. https://doi.org/10.1002/ecy.2142
Fukami T, Bezemer TM, Mortimer SR, Van Der Putten WH (2005) Species divergence and trait convergence in experimental plant community assembly. Ecol Lett 8:1283–1290. https://doi.org/10.1111/j.1461-0248.2005.00829.x
Fundação SOS Mata Atlântica (2014) Atlas dos remanescentes florestais da Mata Atlântica: período 2012–2013. Fundação SOS Mata Atlântica, Instituto Nacional de Pesquisas Espaciais, São Paulo
Hardin G (1960) The competitive exclusion principle. Science 131:1292–1297. https://doi.org/10.1126/science.131.3409.1292
Harvey PH, Pagel MD (1991) The comparative method in evolutionary biology. Oxford University Press, New York
Hérault B (2007) Reconciling niche and neutrality through the emergent group approach. Perspect Plant Ecol Evol Syst 9:71–78. https://doi.org/10.1016/j.ppees.2007.08.001
Hubbell SP, Foster RB (1986) Biology, chance, and history and the structure of tropical rain forest tree communities. In: Diamond J, Case TJ (eds) Community ecology. Harper and Row, New York, pp 314–329
Janzen DH (1970) Herbivores and the number of tree species in tropical forests. Am Nat 104:501–528. https://doi.org/10.1086/282687
Keddy PA (1992) Assembly and response rules: two goals for predictive community ecology. J Veg Sci 3:157–164. https://doi.org/10.2307/3235676
Kraft NJB, Valencia R, Ackerly DD (2008) Functional traits and niche-based tree community assembly in an Amazonian forest. Science 322:580–582. https://doi.org/10.1126/science.1160662
Kraft NJB, Adler PB, Godoy O, James EC, Fuller S, Levine JM (2015) Community assembly, coexistence and the environmental filtering metaphor. Funct Ecol 29:592–599. https://doi.org/10.1111/1365-2435.12345
Kunstler G, Lavergne S, Courbaud B, Thuiller W, Vieilledent G, Zimmermann NE, Kattge J, Coomes DA (2012) Competitive interactions between forest trees are driven by species’ trait hierarchy, not phylogenetic or functional similarity: implications for forest community assembly. Ecol Lett 15:831–840. https://doi.org/10.1111/j.1461-0248.2012.01803.x
Laliberté E, Legendre P (2010) A distance-based framework for measuring functional diversity from multiple traits. Ecology 91:299–305. https://doi.org/10.1890/08-2244.1
Li Y, Shipley B, Price JN, de Dantas VL, Tamme R, Westoby M, Siefert A, Schamp BS, Spasojevic MJ, Jung V, Laughlin DC, Richardson SJ, Le B-PY, Schöb C, Gazol A, Prentice HC, Gross N, Overton J, Cianciaruso MV, Louault F, Kamiyama C, Nakashizuka T, Hikosaka K, Sasaki T, Katabuchi M, Frenette Dussault C, Gaucherand S, Chen N, Vandewalle M, Batalha MA (2018) Habitat filtering determines the functional niche occupancy of plant communities worldwide. J Ecol 106:1001–1009. https://doi.org/10.1111/1365-2745.12802
Liebmann B, Camargo SJ, Seth A, Marengo JA, Carvalho LMV, Allured D, Fu R, Vera CS (2007) Onset and end of the rainy season in South America in observations and the ECHAM 4.5 atmospheric general circulation model. J Clim 20:2037–2050. https://doi.org/10.1175/JCLI4122.1
Lima RAF, Mori DP, Pitta G, Melito MO, Bello C, Magnago LFS, Zwiener VP, Saraiva DD, Marques MCM, de Oliveira AA, Prado PI (2015) How much do we know about the endangered Atlantic Forest? Reviewing nearly 70 years of information on tree community surveys. Biodivers Conserv 24:2135–2148. https://doi.org/10.1007/s10531-015-0953-1
Lopez BE, Burgio KR, Carlucci MB, Palmquist KA, Parada A, Weinberger VP, Hurlbert AH (2016) A new framework for inferring community assembly processes using phylogenetic information, relevant traits and environmental gradients. One Ecosyst 1:1–24. https://doi.org/10.3897/oneeco.1.e9501
Lortie CJ, Brooker RW, Choler P, Kikvidze Z, Michalet R, Pugnaire FI, Callaway RM (2004) Rethinking plant community theory. Oikos 107:433–438. https://doi.org/10.1111/j.0030-1299.2004.13250.x
MacArthur R, Levins R (1967) The limiting similarity, convergence, and divergence of coexisting species. Am Nat 101:377–385. https://doi.org/10.1086/282505
Magallón S, Gómez-Acevedo S, Sánchez-Reyes LL, Hernández-Hernández T (2015) A metacalibrated time-tree documents the early rise of flowering plant phylogenetic diversity. New Phytol 207:437–453. https://doi.org/10.1111/nph.13264
Martins VF, dos Santos Seger GD, Wiegand T, dos Santos FAM (2018) Phylogeny contributes more than site characteristics and traits to the spatial distribution pattern of tropical tree populations. Oikos 127:1368–1379. https://doi.org/10.1111/oik.05142
Mayfield MM, Levine JM (2010) Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol Lett 13:1085–1093. https://doi.org/10.1111/j.1461-0248.2010.01509.x
McDonald R, McKnight M, Weiss D, Selig E, O’Connor M, Violin C, Moody A (2005) Species compositional similarity and ecoregions: Do ecoregion boundaries represent zones of high species turnover? Biol Conserv 126:24–40. https://doi.org/10.1016/j.biocon.2005.05.008
McFadden IR, Bartlett MK, Wiegand T, Turner BL, Sack L, Valencia R, Kraft NJB (2019) Disentangling the functional trait correlates of spatial aggregation in tropical forest trees. Ecology 100:e02591. https://doi.org/10.1002/ecy.2591
Miles L, Grainger A, Phillips O (2004) The impact of global climate change on tropical forest biodiversity in Amazonia. Glob Ecol Biogeogr 13:553–565. https://doi.org/10.1111/j.1466-822X.2004.00105.x
Myers JA, Vellend M, Gardescu S, Marks PL (2004) Seed dispersal by white-tailed deer: implications for long-distance dispersal, invasion, and migration of plants in eastern North America. Oecologia 139:35–44. https://doi.org/10.1007/s00442-003-1474-2
Olson DM, Dinerstein E, Wikramanayake ED, Burgess ND, Powell GVN, Underwood EC, D’Amico JA, Itoua I, Strand HE, Morrison JC, Loucks CJ, Allnutt TF, Ricketts TH, Kura Y, Lamoreux JF, Wettengel WW, Hedao P, Kassem KR (2001) Terrestrial ecoregions of the world: a new map of life on Earth. Bioscience 51:933–938. https://doi.org/10.1641/0006-3568(2001)051[0933:TEOTWA]2.0.CO;2
Ovaskainen O, Abrego N (2020) Joint species distribution modelling: with applications in R, 1st edn. Cambridge University Press, Cambridge
Ovaskainen O, Abrego N, Halme P, Dunson D (2016a) Using latent variable models to identify large networks of species-to-species associations at different spatial scales. Methods Ecol Evol 7:549–555. https://doi.org/10.1111/2041-210X.12501
Ovaskainen O, Roy DB, Fox R, Anderson BJ (2016b) Uncovering hidden spatial structure in species communities with spatially explicit joint species distribution models. Methods Ecol Evol 7:428–436. https://doi.org/10.1111/2041-210X.12502
Ovaskainen O, Tikhonov G, Norberg A, Blanchet FG, Duan L, Dunson D, Roslin T, Abrego N (2017) How to make more out of community data? A conceptual framework and its implementation as models and software. Ecol Lett 20:561–576. https://doi.org/10.1111/ele.12757
Paradis E, Claude J, Strimmer K (2004) APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. https://doi.org/10.1093/bioinformatics/btg412
R Core Team (2019) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/
Ranzato Filardi FL, De Barros F, Baumgratz JFA, Bicudo CEM, Cavalcanti TB, Nadruz Coelho MA, Costa AF, Costa DP, Goldenberg R, Labiak PH, Lanna JM, Leitman P, Lohmann LG, Costa Maia L, Mansano VF, Morim MP, Peralta DF, Pirani JR, Prado J, Roque N, Secco RS, Stehmann JR, Sylvestre LS, Viana PL, Walter BMT, Zimbrão G, Forzza RC et al (2018) Brazilian flora 2020: innovation and collaboration to meet target 1 of the global strategy for plant conservation (GSPC). Rodriguesia 69:1513–1527. https://doi.org/10.1590/2175-7860201869402
Rüger N, Berger U, Hubbell SP, Vieilledent G, Condit R (2011) Growth strategies of tropical tree species: disentangling light and size effects. PLoS ONE 6:e25330. https://doi.org/10.1371/journal.pone.0025330
Sanderson EW, Jaiteh M, Levy MA, Redford KH, Wannebo AV, Woolmer G (2002) The human footprint and the last of the wild. Bioscience 52:891–904. https://doi.org/10.1641/0006-3568(2002)052[0891:THFATL]2.0.CO;2
Segovia RA, Armesto JJ (2015) The Gondwanan legacy in South American biogeography. J Biogeogr 42:209–217. https://doi.org/10.1111/jbi.12459
Seidler TG, Plotkin JB (2006) Seed dispersal and spatial pattern in tropical trees. PLoS Biol 4:2132–2137. https://doi.org/10.1371/journal.pbio.0040344
Silva IA, Batalha MA (2009) Co-occurrence of tree species at fine spatial scale in a woodland cerrado, southeastern Brazil. Plant Ecol 200:277–286. https://doi.org/10.1007/s11258-008-9452-8
Smith JR, Letten AD, Ke PJ, Anderson CB, Hendershot JN, Dhami MK, Dlott GA, Grainger TN, Howard ME, Morrison BML, Routh D, San Juan PA, Mooney HA, Mordecai EA, Crowther TW, Daily GC (2018) A global test of ecoregions. Nat Ecol Evol 2:1889–1896. https://doi.org/10.1038/s41559-018-0709-x
Sommer B, Harrison PL, Beger M, Pandolfi JM (2014) Trait-mediated environmental filtering drives assembly at biogeographic transition zones. Ecology 95:1000–1009. https://doi.org/10.1890/13-1445.1
Swenson NG, Enquist BJ (2009) Opposing assembly mechanisms in a Neotropical dry forest: implications for phylogenetic and functional community ecology. Ecology 90:2161–2170. https://doi.org/10.1890/08-1025.1
Tjur T (2009) Coefficients of determination in logistic regression models—a new proposal: the coefficient of discrimination. Am Stat 63:366–372. https://doi.org/10.1198/tast.2009.08210
van der Valk AG (1981) Succession in wetlands: a Gleasonian approach. Ecology 62:688–696
Warton DI, Blanchet FG, O’Hara RB, Ovaskainen O, Taskinen S, Walker SC, Hui FKC (2015) So many variables: joint modeling in community ecology. Trends Ecol Evol 30:766–779. https://doi.org/10.1016/j.tree.2015.09.007
Webb CO, Ackerly DD, Kembel SW (2008) Phylocom: Software for the analysis of phylogenetic community structure and trait evolution. Bioinformatics 24:2098–2100. https://doi.org/10.1093/bioinformatics/btn358
Wiegand T, Gunatilleke S, Gunatilleke N (2007) Species associations in a heterogeneous Sri Lankan dipterocarp forest. Am Nat 170:E77–E95. https://doi.org/10.1086/521240
Wiens JJ, Donoghue MJ (2004) Historical biogeography, ecology and species richness. Trends Ecol Evol 19:639–644. https://doi.org/10.1016/j.tree.2004.09.011
Wildlife Conservation Society, Center for International Earth Science Information Network (2005) Global human influence index (Geographic), v2 (1995–2004). NASA Socioeconomic Data and Applications Center, Palisades
Wilson JB, Stubbs WJ (2012) Evidence for assembly rules: limiting similarity within a saltmarsh. J Ecol 100:210–221. https://doi.org/10.1111/j.1365-2745.2011.01891.x
Zambrano J, Marchand P, Swenson NG (2017) Local neighbourhood and regional climatic contexts interact to explain tree performance. Proc R Soc B Biol Sci. https://doi.org/10.1098/rspb.2017.0523
Acknowledgements
We thank André Amorim, João L.F. Batista, Marcelo Pansonato, Mariana C. Pardgurschi and Ricardo R. Rodrigues for providing their published or unpublished tree abundance data. We thank Luiz F.S. Magnago for providing additional trait information for several species. We also thank Gregory Pitta, Danilo Mori, Markus Gastauer, Carolina Bello, Natália Targhetta and Geison Castro for helping with the compilation of abundance and trait data from the literature.
Funding
Open access funding provided by University of Helsinki including Helsinki University Central Hospital. This work was partly supported by the Research Council of Norway through its Centres of Excellence Funding Scheme (223257) to EM, VG & OO via Centre for Biodiversity Dynamics, by the Academy of Finland Grants to OO (284601, 273253 and 309581), and by São Paulo Research Foundation (FAPESP) Grant 2013/08722-5 to RAFL. RAFL also received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie Grant Agreement No. 795114. The data collection of some of the major forest plots was supported by grants 99/08515-0 and 99/09635-0 (FAPESP), but many other agencies (including CAPES, CNPq, FAPEMIG, FAPERJ, FAPESC) funded the hundreds of researchers behind the utilized plot data that are stored in the TreeCo database.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by Kyle Palmquist.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Marjakangas, EL., Ovaskainen, O., Abrego, N. et al. Co-occurrences of tropical trees in eastern South America: disentangling abiotic and biotic forces. Plant Ecol 222, 791–806 (2021). https://doi.org/10.1007/s11258-021-01143-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11258-021-01143-3