Abstract
Micromoths can be challenging to identify based on morphology and are frequently omitted in assessments of moth diversity. However, their species richness and biology make them important components of terrestrial ecosystems. In this study we identified 1227 micromoths from a suburban garden at 63° north using DNA barcoding of Malaise trap samples. We recorded 78 different species with the 11 most abundant taxa accounting for 82 % of the catch. The remaining 67 species were represented by fewer than 14 specimens, but the number was often sufficient to provide a good idea of phenology. The larvae of these 78 species all feed on plants common in suburban environments. We show that when facilitated by identifications through DNA barcoding, Malaise traps provide interesting insights into the micromoth communities of suburban environments that might otherwise be overlooked. The use of Malaise traps is beneficial for investigations at high latitudes where light trapping is inefficient for sampling moths due to bright summer nights.
Similar content being viewed by others
Introduction
Urban ecology is a field in constant growth (Cressey 2015). Traditionally, birds, mammals and flowering plants have been considered in urban ecology studies, but butterflies and moths are now often also investigated (Goode 2014). The larger moth species are reasonably well known and easy to identify in temperate regions, but micromoths require more taxonomic expertise and are therefore rarely considered in urban ecology studies. However, extensive investigations of urban moth communities were recently undertaken in Scotland (Lintott et al. 2014) and Michigan, USA (Rice and White 2015).
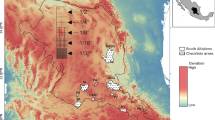
Urbanization is generally supposed to contribute to biodiversity loss (McKinney 2002), but only a few large cities in Norway have an extreme urban structure. In Trondheim (population 190,000), the urban city center is small and most residential or suburban areas consist of houses with gardens surrounded by boreal forest (Fig. 1). Thus, although little is known about insect diversity in the city itself, one would expect the surrounding suburban area to support a rich fauna of terrestrial arthropods.
Lepidoptera are usually collected by light traps or hand nets, because traps that immerse specimens in a preservative, such as ethanol, makes identification using wing patterns difficult. However, identification using standardized molecular markers (i.e. DNA barcoding (Hebert et al. 2003)) alters this situation completely, since ethanol-preserved specimens are perfectly suited for DNA analysis. Flight intercept traps, such as the Malaise trap (Malaise 1937) (Fig. 2), are particularly suitable for collecting micromoths throughout their flight period. Malaise traps are both cost- and time-effective, and have been found to collect a significant portion of the local arthropod fauna despite some bias in taxonomic representation (e.g. Hosking 1979; Noyes 1989). Moreover, Malaise traps that sample the same communities in the same time period produce only insignificant differences in the monitored species composition (Diserud et al. 2013). Thus, Malaise traps are well-suited for comparative studies of the flying arthropod fauna at a large geographical scale.
A major challenge in the analysis of Malaise trap catches is the large number of specimens and the high diversity of taxa present. Sorting and identification by morphology is very time consuming, and it is often difficult to complete for all groups since it requires access to numerous taxonomic specialists. DNA barcoding (Hebert et al. 2003) can overcome this hurdle if a high quality reference library is available to permit species-level matches. Moreover, even if a specific match cannot be made at the time of data acquisition, the deposition of the DNA barcode in BOLD (Ratnasingham and Hebert 2007) allows its assignment to a Barcode Index Number (Ratnasingham and Hebert 2013) and its subsequent assignment to a species when the barcode reference library gains coverage for that taxon. Further efficiency in the analysis of Malaise trap samples can be achieved if combined with high-throughput sequencing technologies, since presorting of the samples then can be reduced to a minimum (Morinière et al. 2016).
DNA barcoding is being used in the Global Malaise Trap Program, a project that seeks to improve understanding of arthropod diversity by using DNA barcoding to identify specimens collected by these traps at sites around the world. The program was initiated in 2012 and has an ever-growing number of participants from all over the world (http://globalmalaise.org). Currently, participants from more than 30 nations from all continents are contributing, building a database which details the temporal and spatial distribution of terrestrial arthropods. Here we report the diversity of micromoths collected by two Malaise traps deployed through the Global Malaise Trap Program and relate these findings to the diversity expected in the temperate suburban area where the traps were deployed. Our results indicate that Malaise traps are effective in collecting micromoths, that DNA barcoding is an effective tool for species-level identifications, and that most micromoth sprecies encountered in this study feed as larvae on plants common in temperate suburban gardens.
Methods
Two Malaise traps (Townes style) were operated in Trondheim, Norway (Figs. 1 and 2) for a 34 week period (March 9 to October 26, 2014) at 63.4050° N 10.3827° E, 70 m.a.s.l. Sample bottles containing 85 % ethanol were changed on a weekly basis, ethanol was refreshed and samples were kept at −20 °C until liquid was decanted immediately before shipment to the Canadian Centre for DNA Barcoding (CCDB) at the University of Guelph. Samples from the two traps collected in a particular week were combined and specimens individually sorted before tissue sampling. Because of the substantial costs of analysis, all 34 weekly samples could not be analyzed. Instead, analysis examined all specimens collected in odd-numbered weeks (1, 3, 5, etc). Small individuals were placed in microplate wells as whole specimens. DNA extraction, PCR and uni-directional (reverse) Sanger sequencing was performed according to the standard protocols used by CCDB. Primers used for Lepidoptera were C_LepFolF and C_LepFolR, cocktails of LCO1490 + LepF1 for 5’end and HCO2198 + LepR1 for 3’end pairing of the standard cytochrome c oxidase subunit 1 (COI) barcode fragment (Folmer et al. 1994; Hebert et al. 2004). All data associated with the Lepidoptera from the project are available under the dataset “Lepidoptera GMP Norway” [DS-LEPGMPN] in the Barcode of Life Data Systems (http://boldsystems.org) (Ratnasingham and Hebert 2007) which can be accessed through the following DOI: dx.doi.org/10.5883/DS-LEPGMPN.
Identifications of all specimens except those belonging to Bucculatrix sp., Dichrorampha sp. Depressaria sp. and the subfamily Olethreutinae were made by comparing the COI sequences with DNA barcodes in BOLD through their Barcode Index Number (BIN) assignments (Ratnasingham and Hebert 2013). The remaining specimens were identified using the identification tool in BOLD, an approach which led to uncertainty in species assignments only for Bucculatrix sp. and Dichrorampha sp. for which the barcode matches were inconclusive. All specimens are deposited as vouchers in the insect collection of the NTNU University Museum, Trondheim, Norway (NTNU-VM). The nomenclature used in this paper follows the BOLD register for family, genus and species levels.
The number of collected specimens for each species in each biweekly period is presented in Table 1.
To evaluate which micromoth species might be local garden residents, a list of food plant preferences for all of the species was compiled and compared with that of the neighborhood vegetation.
Results and discussion
The Malaise trap – an effective trap for micromoths
The biweekly samples from the Malaise traps contained 28,517 terrestrial arthropods and sequences were recovered from 25,177 of them (88.3 % success). Among this total, 1227 were micromoths belonging to 12 superfamilies (Alucitoidea, Choreutoidea, Epermenioidea, Ericranioidea, Gelechioidea, Gracillarioidea, Incurvarioidea, Nepticuloidea, Pterophoroidea, Pyraloidea, Tortricoidea and Yponomeutoidea). The resultant sequence records were assigned to 82 BINs and most of these BINs could be assigned to a species through comparison with the DNA barcode reference library in BOLD. Because members of four species (Agonopterix heracliana, Agonopterix arenella, Anthophila fabricana, Elachista canapennella) were each assigned to two BINs, 78 species were represented in the collections. Several Linnean names were associated with one BIN in two cases so we left their identifications at a genus level (Bucculatrix sp., Dichrorampha sp.). The Malaise traps also collected 111 specimens of macromoths which belonged to 21 species. Because larger moths are better flyers, they are infrequently captured by Malaise traps, but are common in light traps. Although the noctuid Cerapteryx graminis was represented by 67 specimens, the other 20 macromoth species were represented by six or fewer specimens. Because this is less than a quarter of the macromoth species known from light trapping in Trondheim (K. Aagaard, pers. obs.), we do not discuss them further in this study.
Five of the micromoth families collected (Choreutidae, Epermeniidae, Eriocranidae, Glyphipterigidae,Prodoxidae) are rarely found in light traps. Elachistidae are attracted to light, but most species arrive near dawn so they are not trapped if light traps are closed early. The Bucculatricidae is a family of small moths with wingspans less than 9 mm and often overlooked like the species of the species-rich family Gracillariidae. The Gracillariidae are, however, not uncommon in light traps. The two Malaise traps collected ten species belonging to this family, and four (Caloptilia jurateae, C. rufipenella, Euspilapteryx auroguttella and Phyllonorycter emberizaepenella) have never been recorded this far north in Norway. The same result applies to Argyresthia curvella (Argyresthiidae) and Euhyponomeutoides ribesiella (Yponomeutidae). The Nepticulidae, which were represented by four species, is another family with extremely small species in which DNA barcoding can greatly increase our knowledge of taxonomy and distribution (van Nieukerken et al. 2015).
Although Malaise traps collect fewer individuals of Lepidoptera per night than light traps, they are well suited for long-term surveys because they require little maintenance. In a suburban neighborhood, the Malaise trap also attract less attention. They are superior in areas with bright summer nights as light traps are less efficient. For small insects like micromoths, which easily are overlooked in light traps, the conservation of specimens in fluid preservative is perfect, also for molecular analyses. Although Lepidoptera are rarely preserved in fluid in in traditional museum collections, the utility of DNA barcoding and Malaise traps for biodiversity investigations justifies this switch especially since new sequencing technologies allow the DNA barcode analysis of bulk samples (‘metabarcoding’) and thereby reduce the effort needed for manual sorting and identification (Morinière et al. 2016). Moreover, analytical costs are steadily decreasing (e.g. Meier et al. 2016), while the efficiency and accuracy is improving. Thus, given access to bioinformatics expertise, metabarcoding of large Malaise trap samples will be straightforward.
Phenology
The seasonal occurrence data indicate a clear division of species flying in the spring, summer or autumn (Table 1). However, the number of specimens for many species is too low for conclusive seasonal assignments. The short summer at 63° north does not allow many lepidopterans to be multivoltine. Fewer than ten species in the Trondheim region have two generations, and only Plutella xylostella is thought to have three (Bengtsson and Johansson 2011). Twenty species have an early, long flight period as they overwinter as adults and start to fly in March. The Eriocranidae and some Nepticulidae typically fly in April to May, while late summer species that do not overwinter as adults include the Crambidae, Gelechidae and Tortricidae. The large number of both species and specimens collected in midsummer contrast with results from light traps which seldom provide good catches in June and July as day length at this latitude is more than 20 h. Thus, at least for micromoths, Malaise traps are an effective way to census biodiversity at high latitudes. The occurrence of early spring and late autumn species underlines the importance of sampling through the entire snow-free period to obtain a comprehensive understanding of their diversity.
Micromoth diversity in an urban environment
In general, the abundances of Lepidoptera species in collections are influenced by trap efficiency and taxonomic bias as well as by the duration of sampling (annually and diurnally). In addition, the diversity of local species typically depends on the availability of food plants and incidence of immigrant species, reflected by the distances to neighboring biotopes with appropriate host plants. Different aspects of a species’ ecology and morphology impact its migratory ability with body size and population size being positively correlated with dispersal (Nieminen et al. 1999). Moreover, monophagous moths tend to be less migratory than polyphagous and oligophagous taxa irrespective of body size (op. cit.). In our study, the 11 most abundant species were represented by an average of nearly 100 specimens each while 26 species were represented by just a single specimen. The diversity distribution approximates a lognormal abundance distribution which is only partly unveiled (Fig. 3). In our case, at least another 33 species of micromoths await discovery according to Chao’s formula (Chao 1984). The abundance distribution can alternatively be viewed as mixture of a local population with 11 abundant species and twice as many other species that are not local residents. The distribution of the latter species then depends on the biotope diversity of the surrounding area as gardens in urban areas have been found to have a high presence of ‘tourist’ species (Rice and White 2015).
The food plants of most micromoth species recorded in our study are grasses or other common herbs (Fig. 4), a pattern also reported in a similar study from Michigan (Rice and White 2015). Typical garden shrubs or trees (Rosa, Prunus, Rubus, Malus, Ribes) are important hosts, but native trees (Salix, Populus, Betula, Alnus, Ulmus, Fraxinus, Acer) are hosts for even more species of micromoths. Coniferous trees (Picea, Pinus) are common in the more distant neighborhood and host fewer species in this garden biotope. Our list of the most important tree species resembles that of Lintott et al. (2014) although it lacks a few southern species. None of the micromoth species we recorded are associated exclusively with introduced horticultural plants. Thus, we regard the 78 micromoth species as local and regional residents and suspect that the high number of singletons reflect “tourist” species from neighboring habitats where they are more frequent.
The species diversity of micromoths outnumber the larger moths and butterflies by a factor of 2–3 in northern Europe. A higher diversity in ecological niches might allow more species to survive in the suburban landscape, as a habitat too small to sustain large butterfly or moth populations might be sufficient for micromoths. The open residential landscapes of Trondheim with short distances between gardens and woodlands, provides a good prospect for a sustainable, diverse insect fauna in most of the city. This creates functional ecosystems in parks and gardens, providing a closeness with nature that is important for appreciation and conservation of local biodiversity.
We have used the term “micromoth” in this study although it is neither a taxonomic group nor an ecological assemblage. There is a continuous gradient in body size between species of moths with no sharp difference in ecological preferences or function. In a perfect study, all groups of Lepidoptera would be surveyed and all individuals would be identified to a species from samples obtained with non-selective methods, providing representative data on species diversity. Although the present study does not meet this ideal, it shows that DNA barcoding provides interesting information on urban insect diversity by reliably identifying taxonomically challenging moths. As the Barcode of Life reference library approaches completeness for other insect groups, DNA barcoding will provide a very accurate and efficient approach for the species identifications needed for ecological studies and for conservation. Moreover, it will enable species identifications required for citizen science initiatives (e.g. http://lifescanner.net/), providing more consistent and trustworthy identifications for urban nature management.
References
Bengtsson BÅ, Johansson R (2011) Nationalnyckln till Sveriges flora och fauna. Fjärilar: Bronsmalar-rullvinmalar. Lepidoptera: Roeslerstammidae-Lyonetodae. ArtDatabanken, SNU, Uppsala
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scand J Stat 11:265–270
Cressey D (2015) Ecologists embrace their urban side. Nature 524:399–400. doi:10.1038/524399a
Diserud OH, Stur E, Aagaard K (2013) How reliable are Malaise traps for biomonitoring? – A bivariate species abundance model evaluation using alpine Chironomidae (Diptera). Insect Conserv Div 6:561–571. doi:10.1111/icad.12012
Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R (1994) DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I diverse metazoan invertebrates. Mol marine Biol. Biotech 3:294–299
Goode D (2014) Nature in towns and cities. Collins New Naturalist Library, vol 127. William Collins, London.
Hebert PDN, Cywinska A, Ball S, de Waard J (2003) Biological identifications through DNA barcodes. Proc R Soc Lond B 270:313–321. doi:10.1098/rspb.2002.2218
Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W (2004) Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc Natl Acad Sci U S A 101:14812–14817. doi:10.1073/pnas.0406166101
Hosking GP (1979) Trap comparison in the capture of flying Coleoptera. N Z Entomol 7:87–92. doi:10.1080/00779962.1979.9722338
Lintott PR, Bunnefeld N, Fuentes-Montemayor E, Minderman J, Blackmore LM, Goulson D, Park KJ (2014) Moth species richness, abundance and diversity in fragmented urban woodlands: implications for conservation and management strategies. Biodivers Conserv 23:2875–2901. doi:10.1007/s10531-014-0753-z
Malaise R (1937) A new insect-trap. Ent Tidskr 58:148–160
McKinney ML (2002) Urbanization, biodiversity, and conservation: the impacts of urbanization on native species are poorly studied, but educating a highly urbanized human population about these impacts can greatly improve species conservation in all ecosystems. Bioscience 52:883–890. doi:10.1641/0006-3568(2002)052[0883:ubac]2.0.co;2
Meier R, Wong W, Srivathsan A, Foo M (2016) $1 DNA barcodes for reconstructing complex phenomes and finding rare species in specimen-rich samples. Cladistics 32:100–110. doi:10.1111/cla.12115
Morinière J, Cancian de Araujo B, Lam AW, Hausmann A, Balke M, Schmidt S, Hendrich L, Doczkal D, Fartmann B, Arvidsson S, Haszprunar G (2016) Species identification in malaise trap samples by DNA barcoding based on NGS technologies and a scoring matrix. PLoS One 11(5):e0155497. doi:10.1371/journal.pone.0155497
Nieminen M, Rita H, Uuvana P (1999) Body size and migration rate in moths. Ecography 22:697–707. doi:10.1111/j.1600-0587.1999.tb00519.x
Noyes JS (1989) A study of five methods of sampling Hymenoptera (Insecta) in a tropical rainforest, with special reference to the Parasitica. J Nat Hist 23:285–298. doi:10.1080/00222938900770181
Ratnasingham S, Hebert PDN (2007) BOLD: the barcode of life data system (www.barcodinglife.org). Mol Ecol Notes 7:355–364. doi:10.1111/j.1471-8286.2007.01678.x
Ratnasingham S, Hebert PDN (2013) A DNA-based registry for all animal species: the barcode index number (BIN) system. PLoS One 8:e66213. doi:10.1371/journal.pone.0066213
Rice AJ, White PJT (2015) Community patterns in urban moth assemblages. J Lep Soc 69:149–156. doi:10.18473/lepi.69i3.a2
van Nieukerken EL, Doorenweerd C, Mutanen M, Landry J-F, Miller J, deWaard JR (2015) A great inventory of the small: combining BOLD datamining and focused sampling hugely increases knowledge of taxonomy, biology, and distribution of leafmining pygmy moths (Lepidoptera: Nepticulidae. Genome 58. doi:10.1139/gen-2015-0087
Acknowledgments
We thank Elisabeth Stur and Aina Mærk Aspaas for assistance with the Malaise trap collections, and staff at the Centre for Biodiversity Genomics, particularly Claudia Bertrand and Kate Perez, for their roles in coordination of the Global Malaise Program. Thanks also to Marc Daverdin for help with the map, and to Peter White and an anonymous reviewer for helpful comments on the manuscript. DNA barcode data in this publication was generated in collaboration with the Norwegian Barcode of Life Network (NorBOL) funded by the Research Council of Norway and the Norwegian Biodiversity Information Centre. The Global Malaise Trap Program is funded, in part, by the Ontario Ministry of Research and Innovation and the Canada Foundation for Innovation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Aagaard, K., Berggren, K., Hebert, P.D. et al. Investigating suburban micromoth diversity using DNA barcoding of malaise trap samples. Urban Ecosyst 20, 353–361 (2017). https://doi.org/10.1007/s11252-016-0597-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11252-016-0597-2