Abstract
This study aimed to systematically collect and appraise the scientific evidence to answer the research question: What MAP genotypes have been isolated from cattle, sheep, and goats in Latin America and the Caribbean? An electronic search was conducted on three platforms (i.e., OVID®, Web of Science®, SciELO) as well as on the proceedings of the International Colloquium on Paratuberculosis. Inclusion and exclusion criteria were defined a priori and conserved through the systematic process and only articles published in peer-reviewed journals were considered. A total of 26 articles met the definitive inclusion criteria. All were published in English, in 15 different journals, and between 1989 and 2020. The relevant articles reported the use of six different genotyping techniques (i.e., polymerase chain reaction-restriction endonuclease analysis, restriction fragment length polymorphism, type-specific-PCR, mycobacterial interspersed repetitive units-variable number of tandem repeats, multi-locus short sequence repeat, single nucleotide polymorphism) in isolates from seven countries. Genotypes found so far in the region using typing techniques were mainly C type. MIRU-VNTR mostly reported INMV 1, INMV 2, and INMV 11 subtypes, among others. MLSSR reported genotypes from four different countries, reporting nine different subtypes of which 7g–10g–4ggt was the most common for loci 1, 2, and 8, respectively. Regardless the high diversity of techniques used so far to genotype Latin American and Caribbean MAP isolates, the original question of this systematic review has been answered. In addition, a relative genetic similarity between MAP strains recovered from cattle, goats, and sheep unrelatedly of the matrix and geographic origin was identified.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycobacterium avium subsp. paratuberculosis (MAP) is a slow-growing, acid-fast, mycobactin dependent pathogen.
Infection with this bacterium ends up in a chronic inflammation in cattle and other wild and domestic species, referred to as paratuberculosis (PTB) or Johne’s disease (JD) (Clarke 1997; Harris and Barletta 2001). Clinical signs of PTB include diarrhea, weight loss, decreased milk yield, and mortality, leading to important economic losses (McAloon et al. 2016). A major concern on MAP is the effortlessness with which the bacterium is spread, since subclinical or clinically infected animals shed the bacterium in feces and milk, empowering dissemination to susceptible calves, the environment, and dairy by-products for human consumption (Sweeney 1996, 2011; Adhikari et al. 2017; Lorencova et al. 2019). MAP-containing milk is of particular concern because the bacterium has been suggested as a possible link to Crohn’s disease (CD) in humans (Kuenstner et al. 2017).
Most infected ruminants never progress toward the clinical stage, remaining sub-clinically infected by MAP. Such epidemiological particularity possibly involves several factors including host-susceptibility, environment, and pathogenic differences between MAP strains (Ahlstrom et al. 2015; Bryant et al. 2016). MAP genotyping is a valuable tool in epidemiological exploration, leading to a better understanding of MAP-infection dynamics, associated risk factors, transmission behavior, and pathogenesis. Consequently, genotyping-based findings can support the implementation of rational and well-adapted control measures, diagnosis improvement, and vaccine development.
MAP strain K-10 complete genome by Li et al. (2005) was the first step to clarifying the molecular basis of MAP’s physiology and virulence, providing the basis for the development of further generations of diagnostic tests and vaccines.
Besides all investigation efforts worldwide, little is known about MAP genetic diversity among susceptible species and specifically, among different geographical locations —a prerequisite for a well-defined understanding of the epidemiology and biology of this bacterium (Stevenson 2015; Bryant et al. 2016). Therefore, we aimed to systematically collect and appraise the scientific evidence to answer the research question: What MAP genotypes have been isolated from cattle, sheep, and goats in Latin America and the Caribbean?
Materials and methods
This review was designed, performed, and reported in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines (Moher et al. 2009). Through it, we will refer to a citation as a title of an article, detected during the stage of title screening, to an article as an entire paper, publication, or research report.
Search strategy
The process of identifying relevant articles considered a specific research question: What MAP genotypes have been isolated from cattle, sheep, and goats in Latin America and the Caribbean?
The following search procedures were implemented on August 2017 and updated on August 2020. The topic was broken down into components and the search terms use to find relevant citations in databases were [Mycobacterium avium paratuberculosis OR paratuberculosis OR Johne*] AND [ruminant? OR cattle OR bovine? OR cow? OR goat? OR caprine? OR sheep OR ovine? OR livestock] AND [characterization OR molecular OR epidemiol* OR fingerprint* OR phylogen* OR genetic* OR genomic* OR genotype* OR diversi* OR dna OR adn OR isolate* OR strain? OR typ* OR subtype*] AND [vntr? OR variable number tandem repeat? OR miru OR mycobacterial interspersed repetitive unit? OR ssr? OR short sequence repeat? OR mlssr OR ml-ssr OR multi-locus short sequence repeat? OR multilocus short sequence repeat? OR rflp OR restriction fragment length polymorphism OR pfge OR pulsed-field gel electrophoresis OR rda OR redundancy analys* OR ghc OR generalized higher criticism OR dgge OR denaturing gradient gel electrophoresis OR snp* OR single nucleotide polymorphism? OR aflp* OR amplified fragment length polymorphism? OR hrm OR high resolution melt* OR rea OR restriction enzymes analys* OR rapd OR random amplification of polymorphic dna OR mlva OR multiple locus variable-number tandem repeat analys*].
Three search platforms (i.e., OVID®, Web of Science®, SciELO) including seven databases (i.e., MEDLINE, CAB Abstracts, Biological Abstracts®, Embase, Web of Science™ Core Collection, Biological Abstracts®, SciELO Citation Index®) were searched. The proceedings of the 3rd to the 12th International Colloquium on Paratuberculosis (ICP) —held between 1991 and 2014, were available from the platforms explored. The 13th and 14th ICP proceedings (2016 and 2018, respectively) were available at the International Association for Paratuberculosis web site (http://www.paratuberculosis.net/) and were hand-searched for existing published primary studies derived from the abstracts by two authors.
Eligibility screening
The inclusion criteria considered all languages. Nevertheless, only those articles, published in peer-reviewed journals, with an available abstract in English, French, German, Portuguese, or Spanish were initially considered. No publishing year or country limitation was considered. In the case of the ICP proceedings and other abstracts found, studies were included in the search strategy only to identify further citations in peer-reviewed journals.
The first selection of citations was done according to the information contained only in the title. Two of the authors made the selection. The exclusion of the citations was done considering the possibility to answer the research question. The reasons for exclusion were (i) not-related topics (e.g., prevalence, Crohn’s disease, economic impact, Mycobacterium bovis); (ii) not the species of interest (e.g., humans, buffaloes, wildlife, camelids); (iii) not in the region of interest (e.g., Egypt, Europe, India, USA); or (iv) not original articles (e.g., review, book). Duplicated articles were not considered. All citations selected by at least one of the two authors were considered eligible to continue in the process.
The eligible citations were screened using the abstract by two of the authors. Inclusion and exclusion criteria were the same as for the title screening. Conflicting was resolved through consensus between authors and if necessary, by a third author.
The remaining articles were studied using the entire paper by two authors to ensure that they contained relevant information (evidence) to answer the research question. Conflicts were resolved through consensus between authors and if necessary, by a third author.
A kappa coefficient was estimated at each step (i.e., title, abstract, entire paper) as previously reported (Landis and Koch 1977), to measure the inter-rater reliability (and also intra-rater reliability) among the number of yes/no in response to the acceptance of each title in each step and by each author.
Two of the authors searched the reference lists of relevant citations identified by the full-text screening for additional published primary articles (“snowballing”). A second snowballing was done considering the reference list of the articles found by the first snowballing procedure.
Data extraction and descriptive summary
After all available articles were compiled, a descriptive summary was presented by one of the authors, considering bibliographic information, country of isolation/detection, source population, matrix, sample size, genotyping method used (including restriction enzyme, when applies), locus, and results (of typing and subtyping). A second author reviewed the data extraction product.
Epidemiological diagrams
The relation among the BstEII restriction profiles was presented in a dendrogram and the different INMV profiles were presented using probable patterns of evolutionary descent between allelic profiles (clonal relationship), inferred by the goeBURST algorithm (Feil et al. 2004), through the minimum spanning tree (MST). It is assumed that the genetic distance between two INMV patterns is proportional to the difference in the number of repeats at each locus. These relations were established using PHYLOViZ 2 software (http://www.phyloviz.net/; Ribeiro-Gonçalves et al. 2016).
Results
Relevant articles accomplishment
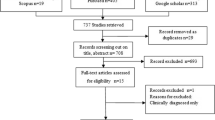
The electronic search found 544 eligible citations (after de-duplication), potentially related to the subject of this systematic review. Manual searching of the 13th and 14th ICP proceedings (2016 and 2018, respectively) provided nine papers. The final number of citations to be furtherly filtered was 553. After reading the titles of the articles, 379 were considered irrelevant (agreed by two authors). The final number of citations based on title screening was 174 (retained by at least one author). After reading the abstracts of the citations, 107 were excluded (by both authors) and 67 original articles remained for the full-text review. Forty-three were excluded because of the criteria already described by title and abstract screening. The snowballing strategy was applied through the reference lists of the 24 definitive articles, and then a second snowballing to the two additional articles kept, leading no new results. Finally, 26 articles met the inclusion criteria of the systematic review (Fig. 1). All were published in English published between July 1989 and August 2020.
The relevant articles reported the use of six different genotyping techniques (i.e., polymerase chain reaction-restriction endonuclease analysis —PCR-REA, restriction fragment length polymorphism —RFLP, type-specific-PCR, mycobacterial interspersed repetitive units-variable number of tandem repeats —MIRU-VNTR, multi-locus short sequence repeat —MLSSR, single nucleotide polymorphism —SNP) in isolates from seven countries of the region of study. One article reported the use of hsp65 PCR-REA, seven of IS1311 PCR-REA, and 11 of IS900-RFLP, using the enzymes BstEII, PstI, HaeIII, HinfI, MseI, PvuII, and BclI. MIRU-VNTR was used in eight articles, considering 17 loci, where the most frequently reported were 1658 (X3), 32, 292, 10, 25, 47, 7, and 3. MLSSR was reported in five articles, mainly considering loci 1, 2, 8, and 9. IS1311 SNP and IS900 type-specific-PCR techniques were reported by one and four articles, respectively. Genotypes found so far in the region using typing techniques were mainly C type. MIRU-VNTR mostly reported INMV 1, INMV 2, and INMV 11 subtypes, among others. MLSSR reported genotypes from four different countries, reporting nine different subtypes of which 7g–10g4ggt was the most common for loci 1, 2, and 8, respectively. Detailed information extracted from the 26 articles included in this systematic review is presented in Tables 1 and 2.
Figure 2 shows epidemiological diagrams obtained and defined over a map of the region of interest with the different MAP animal hosts of study, using the MST diagram for the INMV profiles (A) and a hierarchical clustering diagram, based in BstEII restriction profiles (B).
A goeBURST full minimum spanning tree diagram of INMV profiles, based in the MIRU-VNTRs repeats reported in cattle, sheep, and goats in Latin America and the Caribbean (July 1989–August 2020). B Hierarchical clustering diagram, based in in BstEII restriction profiles reported in cattle, sheep, and goats in Latin America and the Caribbean (July 1989–August 2020)
Discussion
Studies on MAP molecular diversity in Latin America and the Caribbean were reviewed herein, using a systematic approach for the first time on the topic. This report aimed to collect and present all the available data over the available scientific documentation on MAP genotypes isolated from cattle, sheep, and goats in the region of interest so far, expecting to be similar among countries and MAP animal hosts since the countries of the region share some conditions that could define such outcome.
Results obtained in the present review provide important advances in the knowledge of the disease and its etiological agent. Nevertheless, much remains to be accomplished as it has been described in other countries under different productive conditions (Barkema et al. 2017). Such epidemiological evidence could be valuable in the estimation of the transmission of MAP in a herd, among infected facilities from different areas (even countries) or between different animal hosts, and help to improve strategies on the control of the disease.
The number of articles on MAP molecular approaches in the region of interest suggests an increasing trend in PTB research and therefore, an increasing interest in the disease and its negative impacts. In this regard, only 10 articles were published between 1989 and 2005 (during 16 years). As of 2005, with the publication of Li et al. (2005) on the complete genome sequence of MAP, 16 more articles were published in almost 15 years from the seven different countries. It is important to mention that all articles meeting the criteria of the present systematic review were published in English —considered as the language of science, and remarkably, in non-Latin American journals. These results could be founded on the greater visibility that the researchers wanted to give to their findings on the scientific community.
The final number of articles included in the present review (n = 26) is not negligible, but it does represent a significant dilution over time. This could be a result of the interaction of different aspects, including budget limitations (which is a Latin American reality), a low frequency of collaborative research within the region and with other regions, lack of knowledge of the disease plus a low interest in research or screening for it, and because of —in most of the cases, the research front is assumed by the universities, compromising the haste in its development. In addition, since PTB is not a prioritized disease or of mandatory reporting in the region of impact (Whittington et al. 2019), and due to its prolonged incubation period and culturing and testing special features, the expectation of the research product —mainly typing, is not achieved at a short or medium term, since the molecular identification of MAP requires a considerable progress in exploration on the subject, and it is a final step on the research line.
Another aspect that could be analyzed given the results of the present systematic review is related to the evolution in the molecular analysis of MAP, where it can be seen how some countries progressed to techniques that were marking the forefront, while others preserved traditional techniques, which, added to the low number of isolates (due to the factors previously discussed), could explain the limited number of findings for a region as wide and diverse as Latin America and the Caribbean. In this respect another aspect presenting one more limitation is the productive potential or tendency of each country, which explains the lack of information for one country or another on a specific MAP animal host. For example, an interesting question would be if Venezuela does not have MAP molecular findings, since no studies on this topic have been reported in cattle or other susceptible species in this country so far.
According to our results, it can be observed that the Argentinean and Colombian isolates present an observable epidemiological relation, both in the analysis based on MIRU-VNTR profiles and in those of BstEII restriction profiles. For the rest of the isolates, corresponding to the other countries, such similarity was not observed. Any conclusion in this regard should be considered with caution, given the limited number of isolates in some countries, as previously discussed.
The different genotyping techniques presented herein are based on diverse study designs and approaches (e.g., different markers, RFLP-enzymes, matrices, sample size). Therefore, the interpretation derived —related to MAP molecular diversity in the region of interest, is not definitive. The present systematic review is an exploratory study, which allows to visualize the results that, in turn, are not definitive but can contribute with an interesting and well-based overview. In definitive, important limitations are related to the high diversity of matrices and techniques and the low number of studies. For example, Chilean isolates using MIRU-VNTR reported a different set of loci, which does not allow comparisons with the results for the same technique (and even animal species) with those reported in a different country or region of Chile. This is the reason why Chilean findings were not included in Fig. 2.
In definitive, the high variety of methods used so far to genotype Latin American and Caribbean MAP isolates and the limited number of reports makes difficult to analyze the results obtained from the original question of this systematic review. However, we identified a genetic similarity between MAP strains from cattle, goats, and sheep, unrelatedly of the matrix and the geographic origin.
Specific MIRU-VNTR and MLSSR MAP subtypes in the region of study were determined. MIRU-VNTR mostly reported INMV 1, INMV 2, and INMV 11 subtypes. Although comparisons among studies are very difficult because of the use of different analytic loci, it has to be mentioned that MIRU-VNTR profiles 1 (INMV 1) and 2 (INMV 2) have been reported as the most common genotypes found in isolates from European isolates (Stevenson et al. 2009; Douarre et al. 2011; Biet et al. 2012; Imperiale et al. 2017). INMV 11 subtypes seem to be exclusive from the region of study, since no reports on different latitudes have been published so far. MLSSR subtypes were reported from four different countries, leading to nine different subtypes of which 7g–10g–4ggt was the most common one. According to such profile, the subtypes isolated in our study are partially identified by eight codes (i.e., MLSSR 2, MLSSR 5, MLSSR 28, MLSSR 30, MLSSR 31, MLSSR 33, MLSSR 49), according to a web search on http://mac-inmv.tours.inra.fr/macinmv/index.php?p=fa_ident_ssr. Therefore, epidemiological links or relative relation of them with other results in the world is not feasible to establish.
According to the information presented herein, a change to most commonly accepted protocols must be proposed. It is well-known that MAP isolates reveal a restricted diversity and a substitution frequency consistent with a monomorphic pathogen, so it presents a challenge to genotyping (Bryant et al. 2016). Therefore, the leadership of an international entity is proposed to recommend and follow up a conserved classification that allows the tracking of findings all around the world, available from international and freely accessible repositories. This kind of approaches could improve cooperative research among universities, regions, and countries, and of course, would allow to define a differential behavior or dynamic of the study region with respect to the world (which is complicated to the date).
Regarding the methodological process followed in the present systematic review, it included a well-defined protocol based on a clearly defined research question. Two of the authors independently followed selection and results were always by consensus. In a more detailed way, an a priori written protocol (previously reported and endorsed by health-related systematic reviews) based on a clearly stated and delimited research question was followed. A comprehensive search from several databases and sources to identify relevant studies including general-purpose databases, search engines, journals, conference proceedings, book chapters, and books from 1910 (CAB Abstracts) to the date was carried out and the eligibility of included studies by using pre-established and explicit inclusion/exclusion criteria all along the process was assessed. No geographic or temporal constraints were considered, so no biases-related results are thought to be yielded. Two of the authors independently followed selection principles, and results from each searching step were always accomplished by consensus. Agreement measures were reported all along the process to assure reliability of the results. By last, data extracted from the original studies was clearly delineated. Because relevant studies varied in quality and in methodology, one of the authors constructed a matrix of findings, which were furtherly revised by a second author to assure consistency of the information extracted.
We recognize that our SR contains a modest sample of 26 original articles. However, our findings represent the most comprehensive summary of MAP genotypes isolated from cattle, sheep, and goats in Latin America and the Caribbean so far.
Gray literature was not fully considered, because many of these documents are difficult to find and acquire. Therefore, the authors are aware of missing information that could be reporting MAP genotypes in the hosts and regions of interest that may have not been identified. Snowballing (at different levels) strategy was carried out to diminished the effect of such inadvertence, plus to a hand searching of the 14th ICP proceedings, leading to a maximum yield of relevant articles. Bias may have been introduced by the exclusion of non-English, French, German, Portuguese, or Spanish language studies.
In light of the currently available data, systematically reviewing the literature on this subject is difficult as there is considerable variability in study designs and approaches among studies, since individual study designs and genotyping protocols’ characteristics varied considerably among reports. Therefore, it is important to consider the information with the prudence that it deserves and to consider that the outcome of interest (i.e., MAP isolation molecular identity) may vary from case to case. Information from scientific literature is limited, because of studies objectives, designs, and reporting. Authors consider that further studies in this area should follow standardized guidelines when designing and implementing MAP molecular research approaches and reporting their results.
Data availability
Upon request to the corresponding author.
Code availability
Not applicable.
References
Adhikari, S., Tohme, T.C. and Whiley, H., 2017. Investigation of Mycobacterium avium complex (MAC) in Australian commercial milk using qPCR, Journal of Dairy Research, 84(1):89--91
Ahlstrom, C., Barkema, H.W., Stevenson, K., Zadoks, R.N., Biek, R., Kao, R., Trewby, H., Haupstein, D., Kelton, D.F., Fecteau, G., Labrecque, O., Keefe, G.P., McKenna, S.L.B. and De Buck, J., 2015. Limitations of variable number of tandem repeat typing identified through whole genome sequencing of Mycobacterium avium subsp. paratuberculosis on a national and herd level BMC, Genomics, 16, 1–9
Barkema, H.W., Orsel, K., Nielsen, S.S., Koets, A.P., Rutten, V.P.M.G., Bannantine, J.P., Keefe, G.P., Kelton, D.F., Wells, S.J., Whittington, R.J., Mackintosh, C.G., Manning, E.J., Weber, M.F., Heuer, C., Forde, T.L., Ritter, C., Roche, S., Corbett, C.S., Wolf, R., Griebel, P.J., Kastelic, J.P. and De Buck, J., 2017. Knowledge gaps that hamper prevention and control of Mycobacterium avium subspecies paratuberculosis infection, Transboundary and Emerging Diseases, 65(1), 125–148
Biet, F., Sevilla, I.A., Cochard, T., Lefrançois, L.H., Garrido, J.M., Heron, I., Juste, R.A., McLuckie, J., Thibault, V.C. and Supply, P., 2012. Inter-and Intra-subtype genotypic differences that differentiate Mycobacterium avium subspecies paratuberculosis strains, BMC Microbiology, 12, 264
Bryant, J.M., Thibault, V.C., Smith, D.G.E., McLuckie, J., Heron, I., Sevilla, I.A., Biet, F., Harris, S.R., Maskell, D.J., Bentley, S.D., Parkhill, J. and Stevenson, K., 2016. Phylogenomic exploration of the relationships between strains of Mycobacterium avium subspecies paratuberculosis, BMC Genomics, 17, 79
Bull, T.J., Hermon-Taylor, J., Pavlik, I., El-Zaatari, F. and Tizard, M., 2000. Characterization of IS900 loci in Mycobacterium avium subsp. paratuberculosis and development of multiplex PCR typing, Microbiology, 146, 2185–2197
Chávez, G., Trigo, F.J., Svastova, P. and Pavlik, I., 2004. Identificación del polimorfismo genético de aislamientos de Mycobacterium avium subsp. paratuberculosis de caprinos del centro de México, Veterinaria México, 35, 1–7
Clarke, C.J., 1997. The pathology and pathogenesis of paratuberculosis in ruminants and other species, Journal of Comparative Pathology, 116, 217–261
Correa-Valencia, N., Moyano, R.D., Romano, M.I. and Fernández-Silva, J.A., 2020. Molecular typing of Mycobacterium avium subsp. paratuberculosis in dairy cattle herds of Antioquia, Colombia, Revista MVZ Córdoba, 25(3),e1816
de Albuquerque, P.P.F., de Melo, R.P.B., de Farias Brito, M., Bovino, F., de Souza, M.A., Lima, A.M.C., de Oliveira, E.A.A., de Moraes Pereira, H. and Mota, R.A., 2018. First molecular epidemiological study of Mycobacterium avium subsp. paratuberculosis in cattle and buffalo from different regions of Brazil, Tropical Animal Health and Production, 50(8), 1929–1935
de Castro Campos de Souza, M., Lima, M.C., de Freitas Espeschit Braga, I., Schwarz, D.G.G., de Souza Rodrigues, A.P., Sales, E.B., Junior, A.A.F. and Moreira, M.A.S., 2016. Molecular typing of Mycobacterium avium subsp. paratuberculosis (MAP) isolated from dairy goats in Brazil, Small Ruminant Research, 140, 18–21
Douarre, P.E., Cashman, W., Buckley, J., Coffey, A. and O'Mahony, J., 2011. Molecular characterization of Mycobacterium avium subsp. paratuberculosis using multi-locus short sequence repeat (MLSSR) and mycobacterial interspersed repetitive units-variable number tandem repeat (MIRU-VNTR) typing methods, Veterinary Microbiology, 149, 482–487
Favila-Humara, L.C., Chávez-Gris, G.G., Carrillo-Casas, E.M. and Hernández-Castro, R., 2010. Mycobacterium avium subsp. paratuberculosis detection in individual and bulk tank milk samples from bovine herds and caprine flocks, Foodborne Pathogens and Disease, 7(4), 351–355
Feil, E.J., Li, B.C., Aanensen, D.M., Hanage, W.P. and Spratt, B.G., 2004. eBURST: Inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data, Journal of Bacteriology, 186(5),1518–1530
Fernández-Silva, J.A., Abdulmawjood, A., Akineden, Ö. and Bülte, M., 2012. Genotypes of Mycobacterium avium subsp. paratuberculosis from South American countries determined by two methods based on genomic repetitive sequences, Tropical Animal Health and Production, 44, 1123–1126
Fernández-Silva, J.A., Abdulmawjood, A. and Bülte, M., 2011. Diagnosis and molecular characterization of Mycobacterium avium subsp. paratuberculosis from dairy cows in Colombia, Veterinary Medicine International, 2011, 352561
Fiorentino, M.A., Gioffré, A., Cirone, K., Morsella, C., Alonso, B., Delgado, F. and Paolicchi, F., 2012. First isolation of Mycobacterium avium subsp. paratuberculosis in a dairy goat in Argentina: Pathology and molecular characterization, Small Ruminant Research, 108, 133–136
Francois, B., Krishnamoorthy, R. and Elion, J., 1997. Comparative study of Mycobacterium paratuberculosis strains isolated from Crohn’s disease and Johne’s disease using restriction fragment length polymorphism and arbitrarily primed polymerase chain reaction, Epidemiology and Infection, 118, 227–233
Ghadiali, A.H., Strother, M., Naser, S.A. and Manning, E.J.B., 2004. Mycobacterium avium subsp. paratuberculosis strains isolated from crohn’s disease patients and animal species exhibit similar polymorphic locus patterns, Journal of Clinical Microbiology, 42(11), 5345–5348
Giannitti, F., Fraga, M., Caffarena, R.D., Schild, C.O., Banchero, G., Armién, A.G., Travería, G., Marthaler, D., Wells, S.J. and Riet-Correa, F., 2018. Mycobacterium paratuberculosis sheep type strain in Uruguay: Evidence for a wider geographic distribution in South America, The Journal of Infection in Developing Countries, 12(3), 190–195
Gioffré, A., Muñoz, M.C., Alvarado Pinedo, M.F., Vaca, R., Morsella, C., Fiorentino, M.A., Paolicchi, F., Ruybal, P., Zumárraga, M., Travería, G.E. and Romano, M.I., 2015. Molecular typing of Argentinian Mycobacterium avium subsp. paratuberculosis isolates by multiple-locus variable number-tandem repeat analysis, Brazilian Journal of Microbiology, 46, 557–564
Harris, N.B. and Barletta, R.G., 2001. Mycobacterium avium subsp. paratuberculosis in Veterinary Medicine, Clinical Microbiology Reviews, 14(3), 489–512
Imperiale, B.R., Moyano, R.D., Di Giulio, A.B., Romero, M.A., Alvarado Pinedo, M.F., Santangelo, M.P., Travería, G.E., Morcillo, N.S. and Romano, M.I., 2017. Genetic diversity of Mycobacterium avium complex strains isolated in Argentina by MIRU-VNTR, Epidemiology and Infection, 145, 1382–1391
Kuenstner, J.T., Naser, S., Chamberlin, W., Borody, T., Graham, D.Y., McNees, A., Hermon-Taylor, J., Hermon-Taylor, A., Dow, C.T., Thayer, W., Biesecker, J., Collins, M.T., Sechi, L.A., Singh, S.V., Zhang, P., Shafran, I., Weg, S., Telega, G., Rothstein, R., Oken, H., Schimpff, S., Bach, H., Bull, T., Grant, I., Ellingson, J., Dahmen, H., Lipton, J., Gupta, S., Chaubey, K., Singh, M., Agarwal, P., Kumar, A., Misri, J., Sohal, J., Dhama, K., Hemati, Z., Davis, W., Hier, M., Aitken, J., Pierce, E., Parrish, N., Goldberg, N., Kali, M., Bendre, S., Agrawal, G., Baldassano, R., Linn, P., Sweeney, R.W., Fecteau, M., Hofstaedter, C., Potula, R., Timofeeva, O., Geier, S., John, K., Zayanni, N., Malaty, H.M., Kahlenborn, C., Kravitz, A., Bulfon, A., Daskalopoulos, G., Mitchell, H., Neilan, B., Timms, V., Cossu, D., Mameli, G., Angermeier, P., Jelic, T., Goethe, R., Juste, R.A. and Kuenstner, L., 2017. The Consensus from the Mycobacterium avium ssp. paratuberculosis (MAP) Conference, Frontiers in Public Health, 5, 1–5
Landis, J.R. and Koch, G.G., 1977. The measurement of observer agreement for categorical data biometrics, Biometrics, 33(1), 159–174
Li, L., Bannantine, J.P., Zhang, Q., Amonsin, A., May, B.J., Alt, D., Banerji, N., Kanjilal, S. and Kapur, V., 2005. The complete genome sequence of Mycobacterium avium subspecies paratuberculosis Proceedings of the National Academy of Sciences, 102, 12344–12349
Lorencova, A., Babak, V., Kralova, A. and Borilova, G., 2019. Survival of Mycobacterium avium subsp. paratuberculosis in raw fermented sausages during production and storage, Meat Science, 155, 20–26
McAloon, C.G., Whyte, P., More, S.J., Green, M.J., O’Grady, L., Garcia, A. and Doherty, M.L., 2016. The effect of paratuberculosis on milk yield—A systematic review and meta-analysis, Journal of Dairy Science, 99, 1449–1460
Merkal, R.S., 1979. Proposal of ATCC 19698 as the neotype strain of Mycobacterium paratuberculosis, International Journal of Systematic Bacteriology, 29(3), 263–264
Moher, D., Liberati, A., Tetzlaff, J. and Altman, D.G., 2009. Systematic reviews and meta-analyses: The PRISMA Statement, Annulas of Internal Medicine, 151, 264–269
Moreira, A.R., Paolicchi, F., Morsella, C., Zumarraga, M., Cataldi, A., Fabiana, B., Alicia, A., Piet, O., Dick, V.S. and María Isabel, R., 1999. Distribution of IS900 restriction fragment length polymorphism types among animal Mycobacterium avium subsp. paratuberculosis isolates from Argentina and Europe, Veterinary Microbiology, 70, 251–259
Muñoz, M.C., Basulto, G.M., Evangelista, T.R., Navarro, F.M., Vizcarra, V.G. and Valencia, G.L., 2013. Caracterización molecular de Mycobacterium avium subespecie paratuberculosis en bovinos y ovinos de Mexicali, Baja California, México, Revista Mexicana de Ciencias Pecuarias, 4(4), 489–500
Overduin, P., Schouls, L., Roholl, P., Der, A. Van, Mahmmod, N., Herrewegh, A., Van, D., Zanden, A. Van Der and Soolingen, D. Van, 2004. Use of Multilocus Variable-Number Tandem-Repeat analysis for typing Mycobacterium avium subsp paratuberculosis, Journal of Clinical Microbiology, 42(11), 5022–5028
Paolicchi, F.A., Cirone, K.M., Morsella, C.G. and Gioffré, A., 2012. First isolation of Mycobacterium avium subsp paratuberculosis from commercial pasteurized milk in Argentina, Brazilian Journal of Microbiology, 244, 1034–1037
Pavlik, I., Horvathova, A., Dvorska, L., Bartl, J., Svastova, P., du Maine, R. and Rychlik, I., 1999. Standardisation of restriction fragment length polymorphism analysis for Mycobacterium avium subspecies paratuberculosis, Journal of Microbiological Methods, 38, 155–167
Ribeiro-Gonçalves, B., Francisco, A.P., Vaz, C., Ramirez, M. and Carriço, J.A., 2016. PHYLOViZ Online: web-based tool for visualization, phylogenetic inference, analysis and sharing of minimum spanning trees, Nucleic Acids Research, 44(1), 246–251
Romano, M.I., Amadio, A., Bigi, F., Klepp, L., Etchechoury, I., Noto Llana, M., Morsella, C., Paolicchi, F., Pavlik, I., Bartos, M., Leão, S.C. and Cataldi, A., 2005. Further analysis of VNTR and MIRU in the genome of Mycobacterium avium complex, and application to molecular epidemiology of isolates from South America, Veterinary Microbiology, 110, 221–237
Stevenson, K., 2015. Genetic diversity of Mycobacterium avium subspecies paratuberculosis and the influence of strain type on infection and pathogenesis: a review, Veterinary Research, 46(1), 64
Stevenson, K., Alvarez, J., Bakker, D., Biet, F., De Juan, L., Denham, S., Dimareli, Z., Dohmann, K., Gerlach, G.F., Heron, I., Kopecna, M., May, L., Pavlik, I., Sharp, J.M., Thibault, V.C., Willemsen, P., Zadoks, R.N. and Greig, A., 2009. Occurrence of Mycobacterium avium subspecies paratuberculosis across host species and European countries with evidence for transmission between wildlife and domestic ruminants, BMC Microbiology, 9, 1–13
Sweeney, R.W., 2011. Pathogenesis of paratuberculosis. Veterinary Clinics of North America: Food Animal Practice, 27, 537–546
Sweeney, R.W., 1996. Transmission of paratuberculosis. Veterinary Clinics of North America: Food Animale Practice, 12(2), 305–312.
Thibault, V.C., Grayon, M., Boschiroli, M.L., Hubbans, C., Overduin, P., Stevenson, K., Gutierrez, M.C., Supply, P. and Biet, F., 2007. New variable-number tandem-repeat markers for typing Mycobacterium avium subsp. paratuberculosis and M. avium strains: Comparison with IS900 and IS1245 restriction fragment length polymorphism typing, Journal of Clinical Microbiology, 45, 2404–2410
Thibault, V.C., Grayon, M., Boschiroli, M.L., Willery, E., Allix-Béguec, C., Stevenson, K., Biet, F. and Supply, P., 2008. Combined multilocus short-sequence-repeat and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing of Mycobacterium avium subsp. paratuberculosis isolates, Journal of Clinical Microbiology, 46, 4091–4094
Travería, G.E., Zumarraga, M., Etchechoury, I., Romano, M.I., Cataldi, A., Alvarado Pinedo, M.F., Pavlik, I., Pribylova, R. and Romero, J.R., 2013. First identification of Mycobacterium avium paratuberculosis sheep strain in Argentina Brazilian Journal of Microbiology, 44, 897–899
Whipple, D., Kapke, P. and Vary, C., 1990. Identification of restriction fragment length polymorphisms in DNA from Mycobacterium paratuberculosis Journal of Clinical Microbiology, 28, 256–2564
Whipple, D.L., Kapke, P. and Jr., R.A., 1989. Analysis of restriction endonuclease fragment patterns of DNA from Mycobacterium paratuberculosis, Veterinary Microbiology, 19, 189–194
Whittington, R., Donat, K., Weber, M.F., Kelton, D., Nielsen, S.S., Eisenberg, S., Arrigoni, N., Juste, R., Sáez, J.L., Dhand, N., Santi, A., Michel, A., Barkema, H., Kralik, P., Kostoulas, P., Citer, L., Griffin, F., Barwell, R., Moreira, M.A.S., Slana, I., Koehler, H., Singh, S.V., Yoo, H.S., Chávez-Gris, G., Goodridge, A., Ocepek, M., Garrido, J., Stevenson, K., Collins, M., Alonso, B., Cirone, K., Paolicchi, F., Gavey, L., Rahman, M.T., De Marchin, E., Van Praet, W., Bauman, C., Fecteau, G., McKenna, S., Salgado, M., Fernández-Silva, J., Dziedzinska, R., Echeverría, G., Seppänen, J., Thibault, V., Fridriksdottir, V., Derakhshandeh, A., Haghkhah, M., Ruocco, L., Kawaji, S., Momotani, E., Heuer, C., Norton, S., Cadmus, S., Agdestein, A., Kampen, A., Szteyn, J., Frössling, J., Schwan, E., Caldow, G., Strain, S., Carter, M., Wells, S., Munyeme, M., Wolf, R., Gurung, R., Verdugo, C., Fourichon, C., Yamamoto, T., Thapaliya, S., Di Labio, E., Ekgatat, M., Gil, A., Alesandre, A.N., Piaggio, J., Suanes, A. and De Waard, J.H., 2019. Control of paratuberculosis: Who, why and how. A review of 48 countries, BMC Veterinary Research, 15, 198
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Nathalia Correa-Valencia had the idea of the article. The literature search and data analysis, as well as the critical revision of the manuscript were performed by all the authors. The first draft of the manuscript was written by Nathalia Correa-Valencia and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The manuscript does not contain clinical studies or patient data.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Correa-Valencia, N.M., Moyano, R.D., Hernández-Agudelo, M. et al. Mycobacterium avium subsp. paratuberculosis (MAP) molecular diversity in cattle, sheep, and goats from Latin America and the Caribbean: a systematic review. Trop Anim Health Prod 53, 468 (2021). https://doi.org/10.1007/s11250-021-02923-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-021-02923-9