Abstract
In the last years, tremendous progress has been made in the development of CRISPR/Cas-mediated genome editing tools. A number of natural CRISPR/Cas nuclease variants have been characterized. Engineered Cas proteins have been developed to minimize PAM restrictions, off-side effects and temperature sensitivity. Both kinds of enzymes have, by now, been applied widely and efficiently in many plant species to generate either single or multiple mutations at the desired loci by multiplexing. In addition to DSB-induced mutagenesis, specifically designed CRISPR/Cas systems allow more precise gene editing, resulting not only in random mutations but also in predefined changes. Applications in plants include gene targeting by homologous recombination, base editing and, more recently, prime editing. We will evaluate these different technologies for their prospects and practical applicability in plants. In addition, we will discuss a novel application of the Cas9 nuclease in plants, enabling the induction of heritable chromosomal rearrangements, such as inversions and translocations. This technique will make it possible to change genetic linkages in a programmed way and add another level of genome engineering to the toolbox of plant breeding. Also, strategies for tissue culture free genome editing were developed, which might be helpful to overcome the transformation bottlenecks in many crops. All in all, the recent advances of CRISPR/Cas technology will help agriculture to address the challenges of the twenty-first century related to global warming, pollution and the resulting food shortage.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Significance statement
Genome editing tools are evolving rapidly. They enable the generation of single or multiple mutations at the desired loci of the plant genome and thus the targeted removal of undesirable or desirable insertion of beneficial traits in crop plants. This review evaluates various CRISPR/Cas-mediated technologies, including recent applications of prime editing and heritable chromosomal rearrangements, in terms of prospects and applicability. The article thus provides the reader not only with an overview of the latest developments in plant genome editing technologies, but also with decision-making aids for the targeted use of these tools for specific fundamental science questions or applications.
Stefan Schillberg, Fraunhofer Institute for Molecular Biology and Applied Ecology IME, Forckenbeckstrasse 6, 52,074 Aachen, Germany.
Diego Orzaez, Instituto de Biología Molecular y Celular de Plantas (IBMCP), Universitat Politècnica de València-Consejo Superior de Investigaciones Científicas (CSIC), Valencia, Spain.
Introduction
In the middle of the last century, for the first time, plant breeders used the artificial induction of mutations to obtain new varieties. This was achieved by applying genotoxic agents, such as ionizing radiation, which randomly induce multiple genomic double-strand breaks (DSBs) (Stadler 1928). In plants, non-homologous end joining (NHEJ) is the dominant pathway of DSB repair, which often results in mutations at the break site (Puchta 2005). After it had become possible to use site-specific nucleases (SSNs) in multicellular eukaryotes (Puchta et al. 1993), the enzymatic induction of single genomic DSBs came into reach. Different kinds of artificial nucleases, such as Zinc-finger nucleases (ZFNs) and Transcription activator-like effector nucleases (TALENs), have been developed to target DSBs to preselected, unique positions in the genome (Voytas 2013). In principle, SSNs can be used most efficiently for mutagenesis by inducing error-prone NHEJ DSB repair in plants (Salomon and Puchta 1998). At the same time, they can also be used to increase homologous recombination (HR)-mediated gene targeting (GT) by several orders of magnitude (Puchta et al. 1996). In most cases, especially in basic science, reverse genetics approaches aim to generate null mutants to study the function of genes. However, like many other mutations that cause phenotypic changes, beneficial traits in agriculture are often due to gain or change of function. Therefore, the establishment of molecular tools for precise gene modifications is required in agriculture. Although many editing tools have been shown to work efficiently in other organisms, they might be of limited applicability in plants due to cellular or environmental differences. Thus, these tools often have not only to be tested but also adapted before they can be used in plants. Over the last few years, CRISPR/Cas-derived genome engineering technologies—due to their huge potential for medicine and all fields of biology and biotechnology—have been developed extremely fast. The number of approaches that are worth evaluating for their potential application in plants rises continuously. Besides various improvements in base editing (BE), a brand-new design for precise genome editing, prime editing (PE), has been developed. Moreover, CRISPR/Cas induced large chromosomal rearrangement (CR) have been achieved recently, making the breakage as well as the formation of genetic linkages an option for application in crops. As a number of excellent reviews has been published recently on different aspects of CRISPR/Cas applications in plants (Atkins and Voytas 2020; Chen et al. 2019; Schindele et al. 2020; Zhang et al. 2019; Zhu et al. 2020), our review will mainly focus on genome modification tools derived from CRISPR/Cas that were developed in the last two years and successfully applied in plants—from single bases to Mb changes (Fig. 1). We will discuss the potential, but also the limitations, of the respective approaches.
Development of engineered CRISPR/Cas for genome editing
The broadest application of the CRISPR/Cas system in plants is the locus-specific DSB induction into genomic DNA to achieve mutagenesis. There are three CRISPR/Cas variants that are most widely used in plants: Streptococcus pyogenes Cas9 (SpCas9) and Staphylococcus aureus (SaCas9) from the type II system; and Lachnospiraceae bacterium ND2006 (LbCas12a), also called LbCpf1 from the type V system (Jinek et al. 2012; Ran et al. 2015; Zetsche et al. 2015). The use of different orthologs of CRISPR/Cas provides many benefits, such as the expansion of available target sites via different protospacer adjacent motifs (PAMs), the generation of different sizes of insertion-deletion mutations (Indels) or multiplex editing using different nucleases. Moreover, different enzyme activities might be targeted by Cas-mediated DNA binding for more complex manipulation of transcriptional or epigenetic state (Puchta 2016). LbCas12a differs in some intriguing properties from Cas9. Its TTTV PAM can be used to target AT-rich genomic regions. Moreover, LbCas12a frequently causes larger deletions than Cas9 due to its protruding single-strand ends after cleavage. Protein engineering of the Cas proteins further broadens the range of applications. By inducing inactivating mutations in both, the HNH domain and the RuvC domain, dCas9, a protein without nuclease activity but with DNA-binding activity can be obtained. On the other hand, inactivation of only one of the nuclease domains results in nCas9, a protein with nickase activity (Barrangou and Marraffini 2014; Le Cong et al. 2013; Ran et al. 2013). Another important improvement has been the expansion of target sites by changing the PAM requirements through engineering. Two modified SpCas9 nucleases, SpCas9-NG and xCas9, were demonstrated to target to NG PAMs in human cells (Hu et al. 2018; Nishimasu et al. 2018). Application of both enzymes has dramatically increased the numbers of potential target sites in plant genomes (Ge et al. 2019; Hua et al. 2019; Ren et al. 2019; Wang et al. 2019; Zhong et al. 2019). Thus, both, the identification of CRISPR/Cas variants and engineering of known CRISPR/Cas nucleases, have accelerated the speed of CRISPR/Cas technology development. In Table 1, we summarized different variants and orthologs of CRISPR/Cas reported in plants besides the frequently used SpCas9, SaCas9 and LbCas12a.
From the start, plants are difficult subjects for gene editing as they have long reproductive circles and often show a low transformation efficiency. On top of this, the use of sophisticated CRISPR/Cas systems, which had been shown to work efficiently in other organisms, has led to mixed results in plants. Codon-optimization of Cas open reading frames is helpful but no guarantee for high cutting efficiencies. There are many more factors hypothesized to influence editing efficiency. One important example is temperature sensitivity: The comparison of different Cas12a variants in plants, AsCas12a, FnCas12a and LbCas12a, demonstrated that temperature is limiting editing efficiency considerably (Malzahn et al. 2019). Some loci showed drastically enhanced editing efficiencies by LbCas12a when the experiments were performed at 29 °C instead of 22 °C in Arabidopsis. Moreover, it is advisable to test multiple nuclease orthologs to identify the most efficient variants for plant genome editing. An examination of several Cas12b orthologs revealed that AaCas12b was the most efficient one in rice (Ming et al. 2020), whereas other variants, AacCas12b, BvCas12 and BhCas12b v4, were demonstrated to work well in rice, cotton and Arabidopsis (Ming et al. 2020; Wang et al. 2020d; Wu et al. 2020). Another example of mixed results is ScCas9, which was originally reported to have a minimal PAM requirement of NNG (Chatterjee et al. 2018). However, it has been shown in rice that ScCas9 works best with NAG PAMs and its mutagenesis efficiency varied drastically between different loci (Wang et al. 2020c). All these examples show that there can be a huge variation in results when newly developed CRISPR/Cas systems are transferred from other organisms to different plant species. Despite all these experimental difficulties, some CRISPR/Cas improvements were developed in plants first. Based on knowledge obtained from the work on enAsCas12a (Kleinstiver et al. 2019), an improved “temperature-tolerant” LbCas12a (ttLbCas12a) which harbors a D156R point mutation, was developed which had significantly higher editing efficiency in plants in comparison to the wild type enzyme (Huang et al. 2021; Schindele and Puchta 2020). Later on, higher editing efficiencies were also reported in human cells and fungi using this D156R mutated variant (Roux et al. 2020; Tóth et al. 2020). Another engineering strategy is the combination of mutations from different Cas variants. For example, XNG‐Cas9 which carries mutations from both xCas9 and Cas9-NG demonstrated higher editing efficiency than both parent Cas9 variants (Niu et al. 2020), whereas combining modifications of eSpCas9 1.1, SpCas9‐HF1 and HypaCas9 in eHF1‐Cas9 led to a reduction of off-side activity (Liang et al. 2018). Engineered CRISPR/Cas nuclease variants that performed well in the genome editing of plants, have—due to their DNA binding capacity—the potential to efficiently control transcription or epigenetic changes in plants, too. Modification of the guide RNA is another strategy for improving CRISPR/Cas efficiencies and capabilities. The development of the single guide RNA (sgRNA) of SpCas9, a fusion between tracrRNA and crRNA, saves time during the cloning process (Jinek et al. 2012). Further manipulation of the sgRNA provides the possibility of CRISPR/Cas-mediated targeting of different kinds of enzyme activities to specific sites in the genome. The incoporation of MS2 and other kinds of aptamers into the sgRNA for sequence-specific protein binding has been used in plants for transcriptional control, live imaging of chromosomes (Khosravi et al. 2020; Lowder et al. 2018), as well as for base editing. Multiplexing techniques require the simultaneous expression of multiple guide RNAs. This can be achieved via multiple expression cassettes of single sgRNA or multiple sgRNAs can be processed from one transcript using ribozymes. Another strategy consists of tRNA setups. One advantage of these self-processed sgRNAs or crRNAs is that they can not only be transcribed by ubiquitous expressing Pol III promoters but also by many Pol II promoters with cell-type/tissue specific expression or inducible expression (He et al. 2017). Previous reports have also shown that the efficiency of mutagenesis by a tRNA-processed sgRNA is higher than simple sgRNA expression by the same Pol III promoter (Zhang et al. 2018).
Although engineered CRISPR/Cas tools provide ample options for genome editing, potential pitfalls have to be taken into account. A relaxed PAM requirement of SpCas9 might not only result in a higher number of potential target sites, but also in a reduced activity at canonical PAMs in plants (Ge et al. 2019; Hua et al. 2019; Ren et al. 2019; Wang et al. 2019; Zhong et al. 2019). This might be due to the fact that the presence of many more putative PAM sequences in the genome might delay the correct binding to the target (Globyte et al. 2019). This hypothesis might also explain why SaCas9 with a longer PAM of NNGRRT has been more efficient at inducing mutations than SpCas9 or Cas12a in comparable experiments in plant cells (Raitskin et al. 2019; Steinert et al. 2015). Moreover, PAM relaxation might reduce the specificity, which needs to be investigated carefully. A recent report revealed that a PAM-less Cas9 might cleave the gRNA expressing cassette in the T-DNA (Qin et al. 2020b). Small Indels in the gRNA cassette will not immediately destroy the function of the sgRNA, but instead produce a mutated sgRNA, increasing the possibility of off-side targeting. This problem can be solved by changing the sgRNA scaffold sequence but such threat was taken into account when the newest developed Cas9 variants, SpG and SpRY, both with strongly reduced PAM requirement were applied (Walton et al. 2020). However, despite harboring a few potential pitfalls, the development of various CRISPR/Cas tools provides plant biologists with novel tools for genome engineering in plants. An increase in editing efficiency can be achieved by enhancing expression of the Cas9 protein, e.g. by use of a viral replicon, the inclusion of introns in the Cas9 open reading frame, the addition of a translational enhancer, or the suppression of RNA silencing (Mao et al. 2018; Peng et al. 2020; Ramona Grützner et al. 2020; Yu et al. 2020). A tissue-specific induction system allows the analysis of phenotypic consequences of a gene knockout in individual organs or cell types that would be lethal if induced in the germline (Decaestecker et al. 2019; Wang et al. 2020a).
Advances of the base editing technique
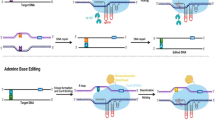
A very important aim of using the CRISPR/Cas system is to achieve precise, predesigned genome modification. As the efficiencies of GT could not be increased to more than a few percent of the transformation events, BE is of special importance for plants. BE is based on the combination of a CRISPR/Cas DNA binding module with a nucleotide base deaminase to achieve one single or a few desired base exchanges. There are two major categories of base editors: cytosine base editors (CBE) that convert C-to-T and adenine base editors (ABE) that convert A-to-G, using a cytosine or adenosine deaminase, respectively. In most cases, nCas9 (D10A) is used in BE to generate a nick in the gRNA binding DNA strand, which enhances the efficiency of the conversion of the nicked strand (Fig. 1) (Bharat et al. 2020).
Multiple studies have shown that both, CBE and ABE, are applicable to different plant species [for reviews see: (Bharat et al. 2020; Zhang et al. 2019; Zhu et al. 2020)]. CBE experiments have been performed with high efficiency in plant cells early on. In contrast, the initial application of ABE in plants resulted in low efficiencies (Hua et al. 2018; Li et al. 2018b; Yan et al. 2018). Recently, by using an optimized adenosine deaminase (Hua et al. 2020b) and increasing PAM accessibility by SpCas9-NG in rice (Hua et al. 2019), improvements of ABE could be achieved. Although CBEs are usually more efficient than ABEs, one advantage of ABEs compared to CBEs is their low off-targeting activity, as demonstrated in the genome of rice and mouse (Jin et al. 2019; Lee et al. 2020). Via whole genome sequencing, it was found that mutations occurred more frequently in transcribed regions, suggesting that single stranded DNAs exposed by transcription are more accessible to cytidine deaminases in a non-specific manner (Jin et al. 2019). Fortunately, the problem of off-target editing of CBE could be solved by the development of two variants, A3Bctd-VHM-BE3 and A3Bctd-KKR-BE3, that have shown high specificity in rice plants at the cost of a reduction in efficiency (Jin et al. 2020).
BEs achieve precise genome editing using their narrow editing window at the target site. Although the ranges differ among various BEs, they are mostly restricted to a 10-bp region (Rees and Liu 2018). As availability of PAM sites is the major restricting factor for access to target sites in the genome, different Cas9 variants can be fused with base deaminases to expand the target range for BE. For example, Cas9-NG, ScCas9 and iSpyMacCas9 were applied to plants, using NG, NAG, and NAAR PAMs, respectively (Hua et al. 2019; Sretenovic et al. 2020; Wang et al. 2020c). Also, the development of SpRY with very low PAM restriction was applicable for BEs in plants (Ren et al. 2021; Walton et al. 2020; Xu et al. 2021). BE by Cas12a or its PAM altered variants has been successfully performed in human cells (Kleinstiver et al. 2019; Li et al. 2018a), but has not been reported in plants to date.
Besides direct fusions of Cas9 proteins and deaminases, base editors were engineered by using aptamer containing sgRNA scaffolds to recruit cognate binding proteins fused to deaminases, and used to generate base edited plants (Li et al. 2020c). Worth mentioning is a different kind of innovative application of ABEs or CBEs, the production of precise short deletions, as demonstrated in rice and wheat (Li et al. 2020c; Wang et al. 2020b). Another intriguing achievement is the dramatically enhanced efficiency of BE by fusion of single stranded DNA binding domain from Rad51 (Zhang et al. 2020), which increases the accessibility of the substrate to the deaminase. However, it should be tested whether this type of fusion will lead to an increase of non-specific mutations in plants. The deaminase itself is also a promising target for improvement, for example, an engineered adenine deaminase (ABE) which carries eight amino acid exchanges in respect to the wild type enzyme was able to enhance 2- to threefold higher editing efficiency in human cells (Gaudelli et al. 2020). A striking new development is a novel kind of BE that has been shown to efficiently induce C-to-G base transversion (Arbab et al. 2020; Kurt et al. 2020). Since the authors did not come up with an explanation for the mechanism, we want to suggest a scenario that is able to explain the phenomenon: after deamination of the C, the resulting U gets eliminated from the DNA by host factors, leaving behind an abasic site. During replication, a translesion polymerase might then incorporate a C opposite to this abasic site by a template-free polymerization. In the next replication cycle, this C serves as template for a G, resulting in the reported transverison (Fig. 2). Intriguingly, occasional C to G transversions were reported before from CBE studies in cotton and rice (Li et al. 2017; Qin et al. 2020a). Therefore, it will only be a matter of time until base editors for transversions will be successfully established in plants, too.
Hypothetical pathway of C-to-G base transversion by using the BE technology. After deamination of the cytidine (C, black), the resulting Uracil base (U, red) is eliminated from the DNA backbone by endogeous uracil-DNA glycosylases (UDGs) or UDG-fused base editors, resulting in an abasic site (a, grey). During DNA repair or replication, translesion polymerase might incorporate a C opposite to this abasic site by a template-free polymerization. Thus a C-to-G transversion is obtained
Improvement of gene targeting
The most ancient form of precise genome editing is GT, using the endogenous HR repair machinery (Paszkowski et al. 1988). Over decades, the extremely low efficiencies of GT experiments hindered any practical applications in plants [for reviews see (Huang and Puchta 2019; Puchta and Fauser 2013)]. A solution to the problem came into reach when it could be demonstrated that site-specific DSBs can enhance GT frequencies by orders of magnitude (Puchta et al. 1996). The application of CRISPR/Cas for the induction of DSBs has become the method of choice for GT, although in most cases site-specific DSBs are repaired by NHEJ, which is the dominant DNA repair pathway in somatic plant cells. Following a number of technical advances during the last years, CRISPR/Cas induced GT could be further improved.
To obtain heritable GT evens, the mutations have to be either transferred to or directly induced in the germline. A successful strategy is the direct DSB induction in egg-cells as has been shown by two different groups in Arabidopsis (Miki et al. 2018; Wolter et al. 2018). One of the approaches consists of an all-in-one construct, including egg-cell promoter driven SaCas9, sgRNA expressing cassette and GT donor. Transformation of this construct into Arabidopsis and analysis of the T2 progeny resulted in individual lines with GT efficiencies of up to 5% (Wolter et al. 2018). Another approach is a two-step process, using sequential transformation for GT (Miki et al. 2018). Using the same egg-cell promoter, a SaCas9 construct was transformed without sgRNA and GT donor. After identifying the transgenic lines with the highest mutation induction capability out of a larger number of candidates, sgRNA cassette and the donor sequence were transformed into these selected lines after propagation to achieve GT. In this case, GT efficiencies of 5–10% could be achieved in two specific lines. The high efficiency of GT might be explained by the fact that in both cases, by involving a large number of Cas9 expressing lines, high expressers could be selected. In addition to nuclease abundance, the nature of the nuclease might help to enhance GT efficiencies. Cas12a cutting differs from Cas9 not only in that it produces 5′overhangs instead of blunt ends or sometimes one nucleotide overhangs (Stephenson et al. 2018), but that the cut is set further away from the seed sequence, which does not tolerate any mismatches during gRNA binding. In case of Cas12a, this might allow repeated cleavage, even if minor NHEJ mutations have been introduced. Several independent recent studies indicated that LbCas12a is indeed able to outperform Cas9 for GT in plants (Li et al. 2020a; van Vu et al. 2020; Wolter and Puchta 2019). GT efficiency could be further increased in Arabidopsis and tobacco using the temperature tolerant version of LbCas12a (Huang et al. 2021; Merker et al. 2020). The system used in Arabidopsis was the “in planta GT” approach. Here the targeting vector is transformed together with the nuclease on a T-DNA resulting in stable lines. By cleavage of target locus as well as the sites flanking the homologous region in the integrated vector, the GT reaction is induced (Fauser et al. 2012). The system was originally set up for Arabidopsis. Recently, it was demonstrated that in planta targeting is also applicable to corn by use of a heat shock promoter-controlled Cas9 expression during tissue culture (Barone et al. 2020). The successful induction of GT in corn may not only be due to the strategy of vector activation. Increasing the growth temperature might improve Cas9 activity as well. A different way to activate the GT vector is to increase the number of donor molecules inside cells. This has been achieved successfully in plant cells using geminiviral replicons (Baltes et al. 2014; Čermák et al. 2015, 2017; Dahan-Meir et al. 2018; Gil-Humanes et al. 2017; van Vu et al. 2020; Wang et al. 2017a). These DNA virus-derived replicons can not only be used to increase the number of GT donors but also the numbers of nuclease expressing cassettes. It has been shown that both approaches contribute individually to the enhancement of GT. The geminiviral replicon in its simplest set up consists of an expression cassette of a replication protein (Rep), in combination with the large intergenic (LIR) and small intergenic regions (SIR), forming a circular replicon. Components from wheat dwarf virus are applicable for GT in cereal cells (Gil-Humanes et al. 2017; Wang et al. 2017a) whereas bean yellow dwarf virus-derived components have been used in Solanaceous species (Baltes et al. 2014; Čermák et al. 2015; Dahan-Meir et al. 2018; van Vu et al. 2020). However, the application of geminiviral replicons for GT has proven to not always be successful. Till now, no report has been published on the production of fertile cereals using this technology. Moreover, efforts to use this technology for heritable GT events in Arabidopsis have failed (Hahn et al. 2018; Pater et al. 2018; Shan et al. 2018). As the Rep protein required for geminiviral replication is involved in the hijacking of the endogenous replication machinery for its DNA synthesis, transfer to the germ line as well as efficient regeneration of single cells to a fertile plant might be limited in presence of viral replicons—at least in a subset of plant species.
A major reason for the low GT efficiency is that plant cells use NHEJ as dominant DSB repair pathway rather than HR. Manipulation of the endogenous DNA repair system or expression of exogenous proteins have been used to enhance GT efficiencies in a number of previous studies. It has been demonstrated that GT efficieny can be increased in Arabidopsis or rice after factors of the classical NHEJ pathway had been knocked out (Endo et al. 2016a; Qi et al. 2013). It will be interesting to test whether suppression of NHEJ by transcriptional repression or Cas13-mediated mRNA degradation [for reviews see: (Mahas et al. 2018; Wolter and Puchta 2018)] will help to enhance GT if combined with the most promising GT approaches. Similarly, overexpression of HR-stimulating proteins could be considered as well. Overexpressing RAD54 from yeast in Arabidopsis egg cells has led to an increased GT efficiency, although no SSN was used in this experiments (Even-Faitelson et al. 2011). However, an approach using the bacterial strand exchange protein RecA did not help to improve DSB induced GT (Reiss et al. 2000).
A novel innovative approach, named tandem repeat-homology-directed repair strategy (TR-HDR), has recently been established. It uses a two-step strategy to obtain also larger predesigned genomic changes like classical GT, but differs in the involved repair pathways (Lu et al. 2020b). First, the authors achieved site-specific insertion of the donor DNA by NHEJ. The double-stranded DNA was chemically modified at both ends in both strands by two nucleotides with phosphorothioate linkages and phosphorylated 5′ ends. These modifications block the degradation of the double-stranded DNA by cellular nucleases but do not inhibit integration. An at least tenfold enhancement of site-specific integration of the extrachromosomal DNA by NHEJ into the DSB site could be achieved. To obtain a seamless predefined modification of the target locus, the authors included a sequence homologous to the target in the template in such a way that a tandem repeat with the desired mutations arose after integration at the genomic locus. In the second step, a DSB was induced by CRISPR/Cas between these tandem repeats. It has been known for a long time that a DSB induced between tandem repeats is repaired efficiently via single strand annealing (SSA) (Siebert and Puchta 2002). This kind of repair, in contrast to classical HR, is as efficient as NHEJ in somatic plant cells. It is non-conservative and leads to the loss of one repeat, including the sequence between the repeats (Puchta 2005). Thus, by combination of a more efficient site-specific integration strategy, based on modified template using NHEJ, induction of SSA and an extra investment of time, the authors were able to increase the efficiency in comparison to classical DSB induced GT which depends on the synthesis strand annealing mechanism of HR (Puchta 1998). A pitfall of this technology might be that due to the large amount of DNA supplied by particle bombardment in the first step, a lot of vector DNA is integrated ectopically at undesired sites of the same genome, which in the end might hinder the production of transgene-free mutants. In this respect, the geminiviral as well as in planta approaches of GT are superior to TR-HDR. In case of in planta GT only a single copy of the vector is available in the cells, excluding simultaneous ectopic integrations. On the other side, geminiviruses seem to have a mechanism to hinder their integration in the host genome, as this might result in virus resistant plants. Till today, integration has not been reported in case of the virus derived replicons.
Prime editing: critical evaluation
As all approaches to achieve precise genome modification discussed until here have their pitfalls either due to low efficiency, strong off-targeting effects, or a small editing window, hopes were flying high in the plant community when an innovative and novel genome editing technique, PE was first introduced. It allows the introduction of different kinds of genomic changes with high efficiency in mammalian cells (Anzalone et al. 2019). The technology relies on a novel CRISPR/Cas9 complex, which is composed of a PE guide RNA (pegRNA) and a protein consisting of a Cas9 nickase (H840A) fused to a reverse transcriptase. The pegRNA can be used as template for reverse transcription. It is a modified sgRNA that contains a primer binding site (PBS) and the sequence to be copied in the genome at its 3′ end (Fig. 3). This pegRNA also acts as sgRNA to define the targeting site of the nickase. The nick is induced in the non-protospacer binding strand and required for the release of a free 3′single stranded DNA end that can be used by the reverse transcriptase as a primer to copy the RNA template sequence into DNA. In this way, the designed modifications can be incorporated seamlessly into the genome. As the change of DNA sequence is limited to only one strand, it is important to safeguard the newly induced mutations from mismatch repair. Therefore, a second nick by another sgRNA is introduced in the complementary DNA strand, which deludes the cellular DNA repair machinery to preserve the sequence in the newly formed double strand in direction of the freshly induced changes. Nicks are present in the newly synthesized but not in the template strand during semiconservative DNA replication. To preserve the genetic information and eliminate mutations, the sequence of the nicked strand will generally be converted by the repair machinery. There are two different strategies: either the nick is introduced in the unedited strand away from the original nick site (called PE3) or directly opposite of the induced change by a sgRNA that only binds to the DNA sequence which had been newly synthesized by the reverse transcription (called PE3b). Thus, use of PE3b ensure that the second nick can only be induced after the first nick was removed during the repair reaction. In case of PE3, paired nicks can arise, which can result in mutagenic Indels, as has been shown in plants (Schiml et al. 2016). In case of PE3b, less unwanted Indel products were recovered than with PE3 (Anzalone et al. 2019).
Prime editing techniques use two nicks. Prime editing (PE) uses a PE guide RNA (pegRNA) and a Cas9 nickase (H840A) fused to a reverse transcriptase to achieve precise genome modifications. After generating the first nick, the pegRNA can be used as the template for reverse transcription, using the primer binding site (PBS) paired with the target sequence. The modified sequence is incorporated only into the nicked strand, resulting in a heteroduplex DNA. A second nick is induced in the other unedited strand to ensure that the modified strand is used for mismatch repair. In PE3 approach, the nick is introduced in the unedited strand away from the first nick site. In PE3b, the second nick is induced only after the modification was incorporated to prevent deleterious outcomes by the presence of paired nicks
A number of independent studies has demonstrated that PE is able to achieve genome modifications in plant protoplasts as well as stable lines, but with low efficiency. These studies were mostly performed in rice but also in tomato, potato and maize (Butt et al. 2020; Hua et al. 2020a; Jiang et al. 2020; Li et al. 2020b; Lin et al. 2020; Lu et al. 2020a; Tang et al. 2020; Veillet et al. 2020a; Xu et al. 2020). Obviously, a couple of different factors might influence the editing efficiency in plants, such as the nature of the reverse transcriptase, thermal conditions, length of the template, length of PBS and the requirement of the second nick. In contrast to initial reports in human cells and yeast, the generation of a second nick in the PE3 and PE3b system did not enhance editing efficiencies in plants (Butt et al. 2020; Lin et al. 2020; Tang et al. 2020). Surprisingly, a plant-specific reverse transcriptase from Cauliflower mosaic virus has shown a lower editing efficiency than a codon-optimized M-MLV (Lin et al. 2020). Editing efficiency rises with temperature: it was twofold higher at 37 °C than 26 °C (Lin et al. 2020). However, no significant differences were observed between incubation at 37 °C and 32 °C (Tang et al. 2020). Another important factor to be considered is what length the reverse transcription template needs to have, to still be copied into the genome with reasonable efficiency. Templates of sizes between 10 and 20-bp have been successfully used, but editing efficiency massively decreases with increasing template length. Major drawbacks of PE in plants are the massive variability of efficiency between loci but also the unwanted Indel production as by-product of the reaction. Depending on the locus, much more unwanted Indels than predesigned template changes were detected. Both kind of mutations were also found in combination. It is still not clear which process steps of PE in plants are the bottlenecks to further increasing editing efficiency. One possibility is that the flap structure between the DNA-RNA complex after reverse transcription might not be processed efficiently in plants. The replacement of the nickase of SpCas9 with the one of SaCas9 has proven to not be successful: only low efficiency or no PE events were observed in the respective experiments (Hua et al. 2020a). However, the result is not too surprising due to the reported low Indel activity of the SaCas9 (N580A) nickase (Friedland et al. 2015).
By now, PE has been demonstrated to work in several plant species. Various parameters might influence its efficiency. Will PE replace BE or GT for predesigned site-specific changes in plant genomes if frequencies get better in the long run? We do not think so. At this moment, PE is simply too inefficient to be considered as an alterative for inducing single base changes in plant genomes. A batch to batch comparison between PE and BE showed mostly lower efficiency of PE than BE, except for a few exceptional loci (Lin et al. 2020). Comparing GT and PE, PE is applicable to the change or insertion of a few nucleotides and GT allows us to integrate predesigned changes in the plant genomes in the range from 10 to 10,000 bp. Nevertheless, the development of PE in plants is still at its infancy. Hopefully, efficiency will rise with effort and time invested similar to what we experienced with the improvements in GT over the years. Therefore, PE, in its specific application window, has definitely the potential to become a promising tool for precise genome editing in crops.
Chromosome engineering
The central aim of breeding is to combine the agronomically best traits of the gene pool of a crop species and to eliminate adverse traits from elite cultivars. Both is possible if the genes coding for the respective favorable traits are not linked to traits with detrimental features. As genes are organized in chromosomes like beads on a string and in most cases are inherited as a unit, this is often not possible, especially if gene loci are physically close to each other. Thus, breeders have a need for technologies to break or stabilize genetic linkages. Obviously, the most direct way to achieve genetic exchange between chromosomes is the induction of a crossover (CO) between homologues in the respective region. In principle, this should be achievable by the use of CRISPR/Cas (Fig. 4). In an outstanding study, the group of Avi Levy demonstrated that targeted DSBs can induce somatic HR using a homologous chromosome as template (Filler Hayut et al. 2017). They developed a selection system in tomato hybrids to identify HR between homologous chromosomes, based on a visual marker gene and single nucleotide polymorphisms. They were able to identify somatic HR events, mostly gene conversions and a putative CO event that, unfortunately, could not be transferred to the next generation. Although this study indicates that “targeted COs” via DSB-induced somatic HR can be accomplished, more work has to be invested to obtain heritable events and estimate the frequency of their occurrence.
Chromosomal rearrangements and their potential application for breeding. The controlled induction of chromosomal rearrangements will enable plant breeders to change the linkage between traits (as illustrated in green, red and black). A novel linkage of two beneficial traits (in green and red) could be created by reciprocal translocations between non-allelic chromosomes (a) or by artificial crossovers (COs) between allelic chromosomes (b). Translocations can also be used to break the linkage between an elite trait (red) and an adverse trait (black) (d, e). Inversions could be used to activate or deactivate meiotic COs in a specific chromosomal region: A CO-dead inverted region could be reversed, making it possible to bring together two beneficial traits from two cultivars (c). The genetic linkage of two beneficial traits on the same chromosome can be fixed by inverting a region containing the respective traits, making this region inaccessible to COs (f)
However, also other kinds of heritable CRs can be achieved by the use of CRISPR/Cas, that might be useful for breeders. In plants, different kinds of CRs occurred not only frequently during evolution, they also can be regarded as one of the driving forces for genome evolution and speciation [for review see: (Schmidt et al. 2019b)]. Moreover, CRs are not rare events but can be observed occasionally during crop breeding or T-DNA transformation (Hu et al. 2017). Inversions of large chromosome regions of F1-hybrids are known to suppress COs (Drouaud et al. 2006; Giraut et al. 2011). Thus, by reverting inversions, genetic linkages between traits that could not be broken before, will become accessible to meiotic COs. An artificially induced inversion might be useful to stabilize a combination of beneficial traits.
Some time ago it could be demonstrated in plants that the simultaneous induction of two DSBs on one chromosome can cause not only a deletion of this region (Siebert and Puchta 2002), but with lower efficiency also its inversion (Schmidt et al. 2019a). Remarkably, a 1.1 Mb natural inversion on chromosome 4 of the Arabidopsis cultivar Col-0 could be recently reversed using SaCas9 for DSB induction (Schmidt et al. 2020). Several independent inversions could be obtained, indicating the efficiency of the used protocol. The resulting revertants were then crossed with an Arabidopsis cultivar devoid of the natural inversion. The authors were able to document COs within the formerly CO-dead region of the genome in the hybrids. Applying this method to crops should allow to restore the CO activity in known inverted regions or to prevent COs between two elite traits (Fig. 4).
In addition, induction of two DSBs on different non-allelic chromosomes can cause reciprocal translocations (Pacher et al. 2007). Whereas cells with dicentric or acentric chromosomes will be lost, reciprocal translocations that maintain the functional chromosome organization should be heritable. Indeed, heritable CRISPR/Cas9-mediated reciprocal translocations were obtained recently by the use of SaCas9 in Arabidopsis (Beying et al. 2020). The authors could induce exchanges in the Mbp range between chromosome 1 and 2, as well as 1 and 5. By cytological and molecular analysis, it could be demonstrated that in some of the translocation lines not a single nucleotide was lost during the exchanges. Thus, reciprocal translocations might turn out to be a novel way to break genetic linkages in a way that was not possible for breeders till now. Similarly, two genes on different chromosomes can be genetically linked if they are placed on the same chromosome in close proximity by an induced translocation (Fig. 4).
Cell culture-free genome editing
One of the limitations of genome editing in plants is the transformation process. The three major transformation methods for plants are Agrobacterium-mediated transformation, biolistic transformation and PEG mediated transformation. Except for Arabidopsis and its close relatives amenable to floral dip transformation, the transformation of most crops requires tissue culture in order to regenerate fertile plants from somatic cells. This step is a time-consuming process and a major bottleneck for many crop plants. It is not only labor-intensive, but can also lead to genetic or epigenetic variation. Very recently, two approaches were developed by the group of Dan Voytas, both of them being able to generate genome edited crops while bypassing the tissue culture step, either by de novo induction of the meristem or by mobile gRNAs to edit meristem (Fig. 5) (Ellison et al. 2020; Maher et al. 2020).
Tissue culture-free plant gene editing. Two innovative approaches to obtain gene edited plants without a tissue-culture process, as demonstrated in tobacco Nicotiana benthamiana. The upper scheme shows that the de novo meristem can be induced by overexpressing growth regulators, such as Wus2, ipt or STM. Novel shoots will be induced which will carry the desired edit in the genome. The shoots could either be propagated for regeneration or might set flowers directly, so that edited seeds should be obtained one way or the other. The lower part of the figure shows that in plants carrying a Cas9 expressing transgene, gene editing can be achieved via a systemic infection with a viral RNA replicon carrying a mobile sgRNA. There is a high probability that shoots and flowers growing after the infection are edited in their genome, resulting in edited seeds in the long run
In tissue-culture, the stimulation of cell growth usually relies on hormone containing medium. The hormones stimulate cell division and keep the resulting callus at a similar cellular stage as the meristematic cells. However, the concentrations needed vary between species and have to be optimized individually. A big step forward has been the increase of tissue-culture efficiency by overexpressing growth factors, such as ipt for dicots (Ebinuma et al. 1997; Smigocki and Owens 1988) or Wus2 for monocots (Lowe et al. 2016, 2018; Nelson-Vasilchik et al. 2018). Recently, a major advance was achieved by Agrobacterium-mediated transformation of a T-DNA with three growth stimulating factors, Wus2, STM and ipt, while editing was induced simultaneously. Thus, genome edited shoots could be regenerated from soil-grown tobacco without a tissue-culture step (Maher et al. 2020). This method has the potential to cut down costs and time of generating gene-edited crops.
Another recent approach with the aim of bypassing tissue-culture has been the utilization of a RNA virus vector from Tobacco rattle virus (Ellison et al. 2020). The virus was manipulated in such a way that it produced a sgRNA fused to the mRNA of FT, a flowering factor that is known to be able to move cell-to-cell, long distances via the phloem and can even cross grafting junctions between species. Transgenic lines of tobacco plants expressing the Cas9 gene were used for virus infection. The authors were not only able to achieve gene editing in the infected leaves, they also found higher editing frequencies in the upper parts of the plants than at the initial infection. Moreover, transfer of the mutation through the germline to the next generation was documented. By using plants with a single copy of the transgene, one might be able to obtain by the segregation of gene-edited progeny that no longer contain a Cas9 expression vector. This kind of RNA-virus mediated delivery of the gRNA should, in principle, also be applicable to BE and PE approaches.
Another method for DNA-free editing of somatic plant cells is the use of the sonchus yellow net rhabdovirus as a vector to express the nuclease Cas9 as well as the gRNA (Ma et al. 2020). Application in crops requires the use of RNA viruses which are able to carry the several kbs of excess genetic information, however the virus in question has only a narrow host range. Unfortunately, most other RNA viruses are not able to carry extra information for coding for such a large protein as Cas9. Nevertheless, as demonstrated by the recent characterization of CasΦ (Pausch et al. 2020), CRISPR/Cas nucleases of much smaller size are becoming available for editing, indicating that this approach might have a promising future as well. Moreover, it might be useful to consider further approaches such as grafting for DNA-free editing. Thus, the next years might see a growing number of options for DNA-free genome editing of plants.
Conclusion
Over the last two years we have seen tremendous progress in the development of CRISPR/Cas-mediated genome editing tools in plants. The establishment of various natural and engineered nucleases has enabled us to target almost any sequence in the genome with high efficiency. We are now able to induce genomic changes from a single base pair to Mbps using technologies from base editing to chromosome engineering. We also have tools at our hands to overcome transformation hurdles in crops. One does not have to be a prophet to predict that also in the next years we will see further major improvements of our genome editing toolbox. This will enable us to supply breeders with the tools they need to breed crops that need less pesticide and are more tolerant to environmental stresses due to global warming. However, challenges remain, especially those beyond technological developments: A scientific based regulation of gene edited organisms everywhere around the globe, an international consensus on how to trade genome edited crops as well as a positive assessment of the technology by the general public will need to be achieved so that mankind will benefit best from these new technologies.
Availability of data and material
Not applicable.
References
Anzalone AV, Randolph PB, Davis JR, Sousa AA, Koblan LW, Levy JM, Chen PJ, Wilson C, Newby GA, Raguram A, Liu DR (2019) Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576:149–157. https://doi.org/10.1038/s41586-019-1711-4
Arbab M, Shen MW, Mok B, Wilson C, Matuszek Ż, Cassa CA, Liu DR (2020) Determinants of base editing outcomes from target library analysis and machine learning. Cell 182:463-480.e30. https://doi.org/10.1016/j.cell.2020.05.037
Atkins PA, Voytas DF (2020) Overcoming bottlenecks in plant gene editing. Curr Opin Plant Biol 54:79–84. https://doi.org/10.1016/j.pbi.2020.01.002
Baltes NJ, Gil-Humanes J, Cermak T, Atkins PA, Voytas DF (2014) DNA replicons for plant genome engineering. Plant Cell 26:151–163. https://doi.org/10.1105/tpc.113.119792
Barone P, Wu E, Lenderts B, Anand A, Gordon-Kamm W, Svitashev S, Kumar S (2020) Efficient gene targeting in maize using inducible CRISPR-Cas9 and marker-free donor template. Mol Plant 13:1219–1227. https://doi.org/10.1016/j.molp.2020.06.008
Barrangou R, Marraffini LA (2014) CRISPR-Cas systems: prokaryotes upgrade to adaptive immunity. Mol Cell 54:234–244. https://doi.org/10.1016/j.molcel.2014.03.011
Begemann MB, Gray BN, January E, Gordon GC, He Y, Liu H, Wu X, Brutnell TP, Mockler TC, Oufattole M (2017) Precise insertion and guided editing of higher plant genomes using Cpf1 CRISPR nucleases. Sci Rep 7:11606. https://doi.org/10.1038/s41598-017-11760-6
Bernabé-Orts JM, Casas-Rodrigo I, Minguet EG, Landolfi V, Garcia-Carpintero V, Gianoglio S, Vázquez-Vilar M, Granell A, Orzaez D (2019) Assessment of Cas12a-mediated gene editing efficiency in plants. Plant Biotechnol J 17:1971–1984. https://doi.org/10.1111/pbi.13113
Beying N, Schmidt C, Pacher M, Houben A, Puchta H (2020) CRISPR-Cas9-mediated induction of heritable chromosomal translocations in Arabidopsis. Nat Plants 6:638–645. https://doi.org/10.1038/s41477-020-0663-x
Bharat SS, Li S, Li J, Yan L, Xia L (2020) Base editing in plants: current status and challenges. Crop J 8:384–395. https://doi.org/10.1016/j.cj.2019.10.002
Butt H, Rao GS, Sedeek K, Aman R, Kamel R, Mahfouz M (2020) Engineering herbicide resistance via prime editing in rice. Plant Biotechnol J. https://doi.org/10.1111/pbi.13399
Čermák T, Baltes NJ, Čegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16:232. https://doi.org/10.1186/s13059-015-0796-9
Čermák T, Curtin SJ, Gil-Humanes J, Čegan R, Kono TJY, Konečná E, Belanto JJ, Starker CG, Mathre JW, Greenstein RL, Voytas DF (2017) A multipurpose toolkit to enable advanced genome engineering in plants. Plant Cell 29:1196–1217. https://doi.org/10.1105/tpc.16.00922
Chatterjee P, Jakimo N, Jacobson JM (2018) Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci Adv 4:eaau0766. https://doi.org/10.1126/sciadv.aau0766
Chen K, Wang Y, Zhang R, Zhang H, Gao C (2019) CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu Rev Plant Biol 70:667–697. https://doi.org/10.1146/annurev-arplant-050718-100049
Dahan-Meir T, Filler-Hayut S, Melamed-Bessudo C, Bocobza S, Czosnek H, Aharoni A, Levy AA (2018) Efficient in planta gene targeting in tomato using geminiviral replicons and the CRISPR/Cas9 system. Plant J 95:5–16. https://doi.org/10.1111/tpj.13932
de Pater S, Klemann BJPM, Hooykaas PJJ (2018) True gene-targeting events by CRISPR/Cas-induced DSB repair of the PPO locus with an ectopically integrated repair template. Sci Rep 8:3338. https://doi.org/10.1038/s41598-018-21697-z
Decaestecker W, Buono RA, Pfeiffer ML, Vangheluwe N, Jourquin J, Karimi M, van Isterdael G, Beeckman T, Nowack MK, Jacobs TB (2019) CRISPR-TSKO: A technique for efficient mstagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 31:2868–2887. https://doi.org/10.1105/tpc.19.00454
Drouaud J, Camilleri C, Bourguignon P-Y, Canaguier A, Bérard A, Vezon D, Giancola S, Brunel D, Colot V, Prum B, Quesneville H, Mézard C (2006) Variation in crossing-over rates across chromosome 4 of Arabidopsis thaliana reveals the presence of meiotic recombination “hot spots.” Genome Res 16:106–114. https://doi.org/10.1101/gr.4319006
Ebinuma H, Sugita K, Matsunaga E, Yamakado M (1997) Selection of marker-free transgenic plants using the isopentenyl transferase gene. Proc Natl Acad Sci USA 94:2117–2121. https://doi.org/10.1073/pnas.94.6.2117
Ellison EE, Nagalakshmi U, Gamo ME, Huang P-J, Dinesh-Kumar S, Voytas DF (2020) Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat Plants 6:620–624. https://doi.org/10.1038/s41477-020-0670-y
Endo M, Mikami M, Toki S (2016a) Biallelic gene targeting in rice. Plant Physiol 170:667–677. https://doi.org/10.1104/pp.15.01663
Endo A, Masafumi M, Kaya H, Toki S (2016b) Efficient targeted mutagenesis of rice and tobacco genomes using Cpf1 from Francisella novicida. Sci Rep 6:38169. https://doi.org/10.1038/srep38169
Endo M, Mikami M, Endo A, Kaya H, Itoh T, Nishimasu H, Nureki O, Toki S (2019) Genome editing in plants by engineered CRISPR-Cas9 recognizing NG PAM. Nat Plants 5:14–17. https://doi.org/10.1038/s41477-018-0321-8
Even-Faitelson L, Samach A, Melamed-Bessudo C, Avivi-Ragolsky N, Levy AA (2011) Localized egg-cell expression of effector proteins for targeted modification of the Arabidopsis genome. Plant J 68:929–937. https://doi.org/10.1111/j.1365-313X.2011.04741.x
Fauser F, Roth N, Pacher M, Ilg G, Sánchez-Fernández R, Biesgen C, Puchta H (2012) In planta gene targeting. Proc Natl Acad Sci USA 109:7535–7540. https://doi.org/10.1073/pnas.1202191109
Filler Hayut S, Melamed Bessudo C, Levy AA (2017) Targeted recombination between homologous chromosomes for precise breeding in tomato. Nat Commun 8:15605. https://doi.org/10.1038/ncomms15605
Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, Kornepati AVR, Sousa A, Collins MA, Jayaram H, Cullen BR, Bumcrot D (2015) Characterization of Staphylococcus aureus Cas9: a smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol 16:257. https://doi.org/10.1186/s13059-015-0817-8
Gaudelli NM, Lam DK, Rees HA, Solá-Esteves NM, Barrera LA, Born DA, Edwards A, Gehrke JM, Lee S-J, Liquori AJ, Murray R, Packer MS, Rinaldi C, Slaymaker IM, Yen J, Young LE, Ciaramella G (2020) Directed evolution of adenine base editors with increased activity and therapeutic application. Nat Biotechnol 38:892–900. https://doi.org/10.1038/s41587-020-0491-6
Ge Z, Zheng L, Zhao Y, Jiang J, Zhang EJ, Liu T, Gu H, Qu L-J (2019) Engineered xCas9 and SpCas9-NG variants broaden PAM recognition sites to generate mutations in Arabidopsis plants. Plant Biotechnol J 17:1865–1867. https://doi.org/10.1111/pbi.13148
Gil-Humanes J, Wang Y, Liang Z, Shan Q, Ozuna CV, Sánchez-León S, Baltes NJ, Starker C, Barro F, Gao C, Voytas DF (2017) High-efficiency gene targeting in hexaploid wheat using DNA replicons and CRISPR/Cas9. Plant J 89:1251–1262. https://doi.org/10.1111/tpj.13446
Giraut L, Falque M, Drouaud J, Pereira L, Martin OC, Mézard C (2011) Genome-wide crossover distribution in Arabidopsis thaliana meiosis reveals sex-specific patterns along chromosomes. PLoS Genet 7:e1002354. https://doi.org/10.1371/journal.pgen.1002354
Globyte V, Lee SH, Bae T, Kim J-S, Joo C (2019) CRISPR/Cas9 searches for a protospacer adjacent motif by lateral diffusion. EMBO J 38. https://doi.org/10.15252/embj.201899466
Hahn F, Eisenhut M, Mantegazza O, Weber APM (2018) Homology-directed repair of a defective Glabrous gene in Arabidopsis with Cas9-based gene targeting. Front Plant Sci 9:424. https://doi.org/10.3389/fpls.2018.00424
He Y, Zhang T, Yang N, Xu M, Yan L, Wang L, Wang R, Zhao Y (2017) Self-cleaving ribozymes enable the production of guide RNAs from unlimited choices of promoters for CRISPR/Cas9 mediated genome editing. J Genet Genomics 44:469–472. https://doi.org/10.1016/j.jgg.2017.08.003
Hu Y, Chen Z, Zhuang C, Huang J (2017) Cascade of chromosomal rearrangements caused by a heterogeneous T-DNA integration supports the double-stranded break repair model for T-DNA integration. Plant J 90:954–965. https://doi.org/10.1111/tpj.13523
Hu JH, Miller SM, Geurts MH, Tang W, Chen L, Sun N, Zeina CM, Gao X, Rees HA, Lin Z, Liu DR (2018) Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 556:57–63. https://doi.org/10.1038/nature26155
Hua K, Tao X, Yuan F, Wang D, Zhu J-K (2018) Precise A·T to G·C base editing in the rice genome. Mol Plant 11:627–630. https://doi.org/10.1016/j.molp.2018.02.007
Hua K, Tao X, Han P, Wang R, Zhu J-K (2019) Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol Plant 12:1003–1014. https://doi.org/10.1016/j.molp.2019.03.009
Hua K, Jiang Y, Tao X, Zhu J-K (2020a) Precision genome engineering in rice using prime editing system. Plant Biotechnol J. https://doi.org/10.1111/pbi.13395
Hua K, Tao X, Liang W, Zhang Z, Gou R, Zhu J-K (2020b) Simplified adenine base editors improve adenine base editing efficiency in rice. Plant Biotechnol J 18:770–778. https://doi.org/10.1111/pbi.13244
Huang T-K, Puchta H (2019) CRISPR/Cas-mediated gene targeting in plants: finally a turn for the better for homologous recombination. Plant Cell Rep 38:443–453. https://doi.org/10.1007/s00299-019-02379-0
Huang TK, Armstrong B, Schindele P, Puchta H (2021) Efficient gene targeting in Nicotiana tabacum using CRISPR/SaCas9 and temperature tolerant LbCas12a. Plant Biotechnol J. https://doi.org/10.1111/pbi.13546
Jiang Y-Y, Chai Y-P, Lu M-H, Han X-L, Lin Q, Zhang Y, Zhang Q, Zhou Y, Wang X-C, Gao C, Chen Q-J (2020) Prime editing efficiently generates W542L and S621I double mutations in two ALS genes in maize. Genome Biol 21:257. https://doi.org/10.1186/s13059-020-02170-5
Jin S, Zong Y, Gao Q, Zhu Z, Wang Y, Qin P, Liang C, Wang D, Qiu J-L, Zhang F, Gao C (2019) Cytosine, but not adenine, base editors induce genome-wide off-target mutations in rice. Science 364:292–295. https://doi.org/10.1126/science.aaw7166
Jin S, Fei H, Zhu Z, Luo Y, Liu J, Gao S, Zhang F, Chen Y-H, Wang Y, Gao C (2020) Rationally designed APOBEC3B Cytosine base editors with improved specificity. Mol Cell 79:728-740.e6. https://doi.org/10.1016/j.molcel.2020.07.005
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337:816–821. https://doi.org/10.1126/science.1225829
Khosravi S, Schindele P, Gladilin E, Dunemann F, Rutten T, Puchta H, Houben A (2020) Application of Aptamers improves CRISPR-based live imaging of plant telomeres. Front Plant Sci 11:1254. https://doi.org/10.3389/fpls.2020.01254
Kim H, Kim S-T, Ryu J, Kang B-C, Kim J-S, Kim S-G (2017) CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat Commun 8:14406. https://doi.org/10.1038/ncomms14406
Kleinstiver BP, Sousa AA, Walton RT, Tak YE, Hsu JY, Clement K, Welch MM, Horng JE, Malagon-Lopez J, Scarfò I, Maus MV, Pinello L, Aryee MJ, Joung JK (2019) Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat Biotechnol 37:276–282. https://doi.org/10.1038/s41587-018-0011-0
Kurt IC, Zhou R, Iyer S, Garcia SP, Miller BR, Langner LM, Grünewald J, Joung JK (2020) CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells. Nat Biotechnol. https://doi.org/10.1038/s41587-020-0609-x
Cong Le, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823. https://doi.org/10.1126/science.1231143
Lee HK, Smith HE, Liu C, Willi M, Hennighausen L (2020) Cytosine base editor 4 but not adenine base editor generates off-target mutations in mouse embryos. Commun Biol 3:19. https://doi.org/10.1038/s42003-019-0745-3
Li J, Sun Y, Du J, Zhao Y, Xia L (2017) Generation of targeted point mutations in rice by a modified CRISPR/Cas9 system. Mol Plant 10:526–529. https://doi.org/10.1016/j.molp.2016.12.001
Li X, Wang Y, Liu Y, Yang B, Wang X, Wei J, Lu Z, Zhang Y, Wu J, Huang X, Yang L, Chen J (2018a) Base editing with a Cpf1-cytidine deaminase fusion. Nat Biotechnol 36:324–327. https://doi.org/10.1038/nbt.4102
Li C, Zong Y, Wang Y, Jin S, Zhang D, Song Q, Zhang R, Gao C (2018b) Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol 19:59. https://doi.org/10.1186/s13059-018-1443-z
Li S, Zhang X, Wang W, Guo X, Wu Z, Du W, Zhao Y, Xia L (2018c) Expanding the scope of CRISPR/Cpf1-mediated genome editing in rice. Mol Plant 11:995–998. https://doi.org/10.1016/j.molp.2018.03.009
Li S, Zhang Y, Xia L, Qi Y (2020a) CRISPR-Cas12a enables efficient biallelic gene targeting in rice. Plant Biotechnol J 18:1351–1353. https://doi.org/10.1111/pbi.13295
Li H, Li J, Chen J, Yan L, Xia L (2020b) Precise modifications of both exogenous and endogenous genes in rice by prime editing. Mol Plant 13:671–674. https://doi.org/10.1016/j.molp.2020.03.011
Li C, Zong Y, Jin S, Zhu H, Lin D, Li S, Qiu J-L, Wang Y, Gao C (2020c) SWISS: multiplexed orthogonal genome editing in plants with a Cas9 nickase and engineered CRISPR RNA scaffolds. Genome Biol 21:141. https://doi.org/10.1186/s13059-020-02051-x
Li C, Zhang R, Meng X, Chen S, Zong Y, Lu C, Qiu J-L, Chen Y-H, Li J, Gao C (2020d) Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat Biotechnol 38:875–882. https://doi.org/10.1038/s41587-019-0393-7
Liang Z, Chen K, Yan Y, Zhang Y, Gao C (2018) Genotyping genome-edited mutations in plants using CRISPR ribonucleoprotein complexes. Plant Biotechnol J 16:2053–2062. https://doi.org/10.1111/pbi.12938
Lin Q, Zong Y, Xue C, Wang S, Jin S, Zhu Z, Wang Y, Anzalone AV, Raguram A, Doman JL, Liu DR, Gao C (2020) Prime genome editing in rice and wheat. Nat Biotechnol 38:582–585. https://doi.org/10.1038/s41587-020-0455-x
Lowder LG, Zhou J, Zhang Y, Malzahn A, Zhong Z, Hsieh T-F, Voytas DF, Zhang Y, Qi Y (2018) Robust transcriptional activation in plants using multiplexed CRISPR-Act2.0 and mTALE-Act systems. Mol Plant 11:245–256. https://doi.org/10.1016/j.molp.2017.11.010
Lowe K, Wu E, Wang N, Hoerster G, Hastings C, Cho M-J, Scelonge C, Lenderts B, Chamberlin M, Cushatt J, Wang L, Ryan L, Khan T, Chow-Yiu J, Hua W, Yu M, Banh J, Bao Z, Brink K, Igo E, Rudrappa B, Shamseer PM, Bruce W, Newman L, Shen B, Zheng P, Bidney D, Falco C, Register J, Zhao Z-Y, Xu D, Jones T, Gordon-Kamm W (2016) Morphogenic regulators baby boom and Wuschel improve monocot transformation. Plant Cell 28:1998–2015. https://doi.org/10.1105/tpc.16.00124
Lowe K, La Rota M, Hoerster G, Hastings C, Wang N, Chamberlin M, Wu E, Jones T, Gordon-Kamm W (2018) Rapid genotype “independent” Zea mays L. (maize) transformation via direct somatic embryogenesis. Vitro Cell Dev Biol -Plant 54:240–252. https://doi.org/10.1007/s11627-018-9905-2
Lu Y, Tian Y, Shen R, Yao Q, Zhong D, Zhang X, Zhu J-K (2020a) Precise genome modification in tomato using an improved prime editing system. Plant Biotechnol J. https://doi.org/10.1111/pbi.13497
Lu Y, Tian Y, Shen R, Yao Q, Wang M, Chen M, Dong J, Zhang T, Li F, Lei M, Zhu J-K (2020b) Targeted, efficient sequence insertion and replacement in rice. Nat Biotechnol 38:1402–1407. https://doi.org/10.1038/s41587-020-0581-5
Ma X, Zhang X, Liu H, Li Z (2020) Highly efficient DNA-free plant genome editing using virally delivered CRISPR-Cas9. Nat Plants 6:773–779. https://doi.org/10.1038/s41477-020-0704-5
Mahas A, Neal Stewart C, Mahfouz MM (2018) Harnessing CRISPR/Cas systems for programmable transcriptional and post-transcriptional regulation. Biotechnol Adv 36:295–310. https://doi.org/10.1016/j.biotechadv.2017.11.008
Maher MF, Nasti RA, Vollbrecht M, Starker CG, Clark MD, Voytas DF (2020) Plant gene editing through de novo induction of meristems. Nat Biotechnol 38:84–89. https://doi.org/10.1038/s41587-019-0337-2
Malzahn AA, Tang X, Lee K, Ren Q, Sretenovic S, Zhang Y, Chen H, Kang M, Bao Y, Zheng X, Deng K, Zhang T, Salcedo V, Wang K, Zhang Y, Qi Y (2019) Application of CRISPR-Cas12a temperature sensitivity for improved genome editing in rice, maize, and Arabidopsis. BMC Biol 17:9. https://doi.org/10.1186/s12915-019-0629-5
Mao Y, Yang X, Zhou Y, Zhang Z, Botella JR, Zhu J-K (2018) Manipulating plant RNA-silencing pathways to improve the gene editing efficiency of CRISPR/Cas9 systems. Genome Biol 19:149. https://doi.org/10.1186/s13059-018-1529-7
Merker L, Schindele P, Huang T-K, Wolter F, Puchta H (2020) Enhancing in planta gene targeting efficiencies in Arabidopsis using temperature-tolerant CRISPR/LbCas12a. Plant Biotechnol J 18:2382–2384. https://doi.org/10.1111/pbi.13426
Miki D, Zhang W, Zeng W, Feng Z, Zhu J-K (2018) CRISPR/Cas9-mediated gene targeting in Arabidopsis using sequential transformation. Nat Commun 9:1–9. https://doi.org/10.1038/s41467-018-04416-0
Ming M, Ren Q, Pan C, He Y, Zhang Y, Liu S, Zhong Z, Wang J, Malzahn AA, Wu J, Zheng X, Zhang Y, Qi Y (2020) CRISPR-Cas12b enables efficient plant genome engineering. Nat Plants 6:202–208. https://doi.org/10.1038/s41477-020-0614-6
Negishi K, Kaya H, Abe K, Hara N, Saika H, Toki S (2019) An adenine base editor with expanded targeting scope using SpCas9-NGv1 in rice. Plant Biotechnol J 17:1476–1478. https://doi.org/10.1111/pbi.13120
Nelson-Vasilchik K, Hague J, Mookkan M, Zhang ZJ, Kausch A (2018) Transformation of recalcitrant Sorghum varieties facilitated by baby boom and Wuschel2. Curr Protoc Plant Biol 3:e20076. https://doi.org/10.1002/cppb.20076
Nishimasu H, Shi X, Ishiguro S, Gao L, Hirano S, Okazaki S, Noda T, Abudayyeh OO, Gootenberg JS, Mori H, Oura S, Holmes B, Tanaka M, Seki M, Hirano H, Aburatani H, Ishitani R, Ikawa M, Yachie N, Zhang F, Nureki O (2018) Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 361:1259–1262. https://doi.org/10.1126/science.aas9129
Niu Q, Wu S, Li Y, Yang X, Liu P, Xu Y, Lang Z (2020) Expanding the scope of CRISPR/Cas9-mediated genome editing in plants using an xCas9 and Cas9-NG hybrid. J Integr Plant Biol 62:398–402. https://doi.org/10.1111/jipb.12886
Pacher M, Schmidt-Puchta W, Puchta H (2007) Two unlinked double-strand breaks can induce reciprocal exchanges in plant genomes via homologous recombination and nonhomologous end joining. Genetics 175:21–29. https://doi.org/10.1534/genetics.106.065185
Paszkowski J, Baur M, Bogucki A, Potrykus I (1988) Gene targeting in plants. EMBO J 7:4021–4026. https://doi.org/10.1002/j.1460-2075.1988.tb03295.x
Pausch P, Al-Shayeb B, Bisom-Rapp E, Tsuchida CA, Li Z, Cress BF, Knott GJ, Jacobsen SE, Banfield JF, Doudna JA (2020) CRISPR-CasΦ from huge phages is a hypercompact genome editor. Science 369:333–337. https://doi.org/10.1126/science.abb1400
Peng F, Zhang W, Zeng W, Zhu J-K, Miki D (2020) Gene targeting in Arabidopsis via an all-in-one strategy that uses a translational enhancer to aid Cas9 expression. Plant Biotechnol J 18:892–894. https://doi.org/10.1111/pbi.13265
Puchta H, Dujon B, Hohn B (1993) Homologous recombination in plant cells is enhanced by in vivo induction of double strand breaks into DNA by a site-specific endonuclease. Nucleic Acids Res 21:5034–5040. https://doi.org/10.1093/nar/21.22.5034
Puchta H, Dujon B, Hohn B (1996) Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc Natl Acad Sci USA 93:5055–5060. https://doi.org/10.1073/pnas.93.10.5055
Puchta H (1998) Repair of genomic double-strand breaks in somatic plant cells by one-sided invasion of homologous sequences. Plant J 13:331–339. https://doi.org/10.1046/j.1365-313X.1998.00035.x
Puchta H (2005) The repair of double-strand breaks in plants: mechanisms and consequences for genome evolution. J Exp Bot 56:1–14. https://doi.org/10.1093/jxb/eri025
Puchta H (2016) Using CRISPR/Cas in three dimensions: towards synthetic plant genomes, transcriptomes and epigenomes. Plant J 87:5–15. https://doi.org/10.1111/tpj.13100
Puchta H, Fauser F (2013) Gene targeting in plants: 25 years later. Int J Dev Biol 57:629–637. https://doi.org/10.1387/ijdb.130194hp
Qi Y, Zhang Y, Zhang F, Baller JA, Cleland SC, Ryu Y, Starker CG, Voytas DF (2013) Increasing frequencies of site-specific mutagenesis and gene targeting in Arabidopsis by manipulating DNA repair pathways. Genome Res 23:547–554. https://doi.org/10.1101/gr.145557.112
Qin L, Li J, Wang Q, Xu Z, Sun L, Alariqi M, Manghwar H, Wang G, Li B, Ding X, Rui H, Huang H, Lu T, Lindsey K, Daniell H, Zhang X, Jin S (2020a) High-efficient and precise base editing of C•G to T•A in the allotetraploid cotton (Gossypium hirsutum) genome using a modified CRISPR/Cas9 system. Plant Biotechnol J 18:45–56. https://doi.org/10.1111/pbi.13168
Qin R, Li J, Liu X, Xu R, Yang J, Wei P (2020b) SpCas9-NG self-targets the sgRNA sequence in plant genome editing. Nat Plants 6:197–201. https://doi.org/10.1038/s41477-020-0603-9
Raitskin O, Schudoma C, West A, Patron NJ (2019) Comparison of efficiency and specificity of CRISPR-associated (Cas) nucleases in plants: An expanded toolkit for precision genome engineering. PLoS ONE 14:e0211598. https://doi.org/10.1371/journal.pone.0211598
Ramona Grützner, Patrick Martin, Claudia Horn, Samuel Mortensen, Erin J. Cram, Carolyn W.T. Lee-Parsons, Johannes Stuttmann, Sylvestre Marillonnet (2020) High-efficiency genome editing in plants mediated by a Cas9 gene containing multiple introns. Plant Commun:100135. https://doi.org/10.1016/j.xplc.2020.100135
Ran FA, Hsu PD, Lin C-Y, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F (2013) Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell 154:1380–1389. https://doi.org/10.1016/j.cell.2013.08.021
Ran FA, Cong Le, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, Koonin EV, Sharp PA, Zhang F (2015) In vivo genome editing using Staphylococcus aureus Cas9. Nature 520:186–191. https://doi.org/10.1038/nature14299
Rees HA, Liu DR (2018) Base editing: precision chemistry on the genome and transcriptome of living cells. Nat Rev Genet 19:770–788. https://doi.org/10.1038/s41576-018-0059-1
Reiss B, Schubert I, Köpchen K, Wendeler E, Schell J, Puchta H (2000) RecA stimulates sister chromatid exchange and the fidelity of double-strand break repair, but not gene targeting, in plants transformed by Agrobacterium. Proc Natl Acad Sci USA 97:3358–3363. https://doi.org/10.1073/pnas.97.7.3358
Ren B, Liu L, Li S, Kuang Y, Wang J, Zhang D, Zhou X, Lin H, Zhou H (2019) Cas9-NG greatly expands the targeting scope of the genome-editing toolkit by recognizing NG and other atypical PAMs in rice. Mol Plant 12:1015–1026. https://doi.org/10.1016/j.molp.2019.03.010
Ren Q, Sretenovic S, Liu S, Tang X, Huang L, He Y, Liu L, Guo Y, Zhong Z, Liu G, Cheng Y, Zheng X, Pan C, Yin D, Zhang Y, Li W, Qi L, Li C, Qi Y, Zhang Y (2021) PAM-less plant genome editing using a CRISPR-SpRY toolbox. Nat Plants. https://doi.org/10.1038/s41477-020-00827-4
Roux I, Woodcraft C, Hu J, Wolters R, Gilchrist CLM, Chooi Y-H (2020) CRISPR-mediated activation of biosynthetic gene clusters for bioactive molecule discovery in filamentous fungi. ACS Synth Biol 9:1843–1854. https://doi.org/10.1021/acssynbio.0c00197
Salomon S, Puchta H (1998) Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J 17:6086–6095. https://doi.org/10.1093/emboj/17.20.6086
Schiml S, Fauser F, Puchta H (2016) Repair of adjacent single-strand breaks is often accompanied by the formation of tandem sequence duplications in plant genomes. Proc Natl Acad Sci USA 113:7266–7271. https://doi.org/10.1073/pnas.1603823113
Schindele A, Dorn A, Puchta H (2020) CRISPR/Cas brings plant biology and breeding into the fast lane. Curr Opin Biotechnol 61:7–14. https://doi.org/10.1016/j.copbio.2019.08.006
Schindele P, Puchta H (2020) Engineering CRISPR/LbCas12a for highly efficient, temperature-tolerant plant gene editing. Plant Biotechnol J 18:1118–1120. https://doi.org/10.1111/pbi.13275
Schmidt C, Pacher M, Puchta H (2019a) Efficient induction of heritable inversions in plant genomes using the CRISPR/Cas system. Plant J 98:577–589. https://doi.org/10.1111/tpj.14322
Schmidt C, Schindele P, Puchta H (2019b) From gene editing to genome engineering: restructuring plant chromosomes via CRISPR/Cas. aBIOTECH 1:21–31. https://doi.org/10.1007/s42994-019-00002-0
Schmidt C, Fransz P, Rönspies M, Dreissig S, Fuchs J, Heckmann S, Houben A, Puchta H (2020) Changing local recombination patterns in Arabidopsis by CRISPR/Cas mediated chromosome engineering. Nat Commun 11:4418. https://doi.org/10.1038/s41467-020-18277-z
Shan Q, Baltes NJ, Atkins P, Kirkland ER, Zhang Y, Baller JA, Lowder LG, Malzahn AA, Haugner JC, Seelig B, Voytas DF, Qi Y (2018) ZFN, TALEN and CRISPR-Cas9 mediated homology directed gene insertion in Arabidopsis: a disconnect between somatic and germinal cells. J Genet Genomics 45:681–684. https://doi.org/10.1016/j.jgg.2018.07.011
Siebert R, Puchta H (2002) Efficient repair of genomic double-strand breaks by homologous recombination between directly repeated sequences in the plant genome. Plant Cell 14:1121–1131. https://doi.org/10.1105/tpc.001727
Smigocki AC, Owens LD (1988) Cytokinin gene fused with a strong promoter enhances shoot organogenesis and zeatin levels in transformed plant cells. Proc Natl Acad Sci USA 85:5131–5135. https://doi.org/10.1073/pnas.85.14.5131
Sretenovic S, Yin D, Levav A, Selengut JD, Mount SM, Qi Y (2020) Expanding plant genome-editing scope by an engineered iSpyMacCas9 system that targets A-Rich PAM sequences. Plant Commun:100101. https://doi.org/10.1016/j.xplc.2020.100101
Stadler LJ (1928) Genetic effects of X-rays in maize. Proc Natl Acad Sci 14:69–75. https://doi.org/10.1073/pnas.14.1.69
Steinert J, Schiml S, Fauser F, Puchta H (2015) Highly efficient heritable plant genome engineering using Cas9 orthologues from Streptococcus thermophilus and Staphylococcus aureus. Plant J 84:1295–1305. https://doi.org/10.1111/tpj.13078
Stephenson AA, Raper AT, Suo Z (2018) Bidirectional degradation of DNA cleavage products catalyzed by CRISPR/Cas9. J Am Chem Soc 140:3743–3750. https://doi.org/10.1021/jacs.7b13050
Tang X, Lowder LG, Zhang T, Malzahn AA, Zheng X, Voytas DF, Zhong Z, Chen Y, Ren Q, Li Q, Kirkland ER, Zhang Y, Qi Y (2017) A CRISPR-Cpf1 system for efficient genome editing and transcriptional repression in plants. Nat Plants 3:17018. https://doi.org/10.1038/nplants.2017.18
Tang X, Sretenovic S, Ren Q, Jia X, Li M, Fan T, Yin D, Xiang S, Guo Y, Liu L, Zheng X, Qi Y, Zhang Y (2020) Plant prime editors enable precise gene editing in rice cells. Mol Plant 13:667–670. https://doi.org/10.1016/j.molp.2020.03.010
Tóth E, Varga É, Kulcsár PI, Kocsis-Jutka V, Krausz SL, Nyeste A, Welker Z, Huszár K, Ligeti Z, Tálas A, Welker E (2020) Improved LbCas12a variants with altered PAM specificities further broaden the genome targeting range of Cas12a nucleases. Nucleic Acids Res 48:3722–3733. https://doi.org/10.1093/nar/gkaa110
van Vu T, Sivankalyani V, Kim E-J, Doan DTH, Tran MT, Kim J, Sung YW, Park M, Kang YJ, Kim J-Y (2020) Highly efficient homology-directed repair using CRISPR/Cpf1-geminiviral replicon in tomato. Plant Biotechnol J 18:2133–2143. https://doi.org/10.1111/pbi.13373
Veillet F, Kermarrec M-P, Chauvin L, Guyon-Debast A, Chauvin J-E, Gallois J-L, Nogué F (2020a) Prime editing is achievable in the tetraploid potato, but needs improvement. bioRxiv. https://doi.org/10.1101/2020.06.18.159111
Veillet F, Perrot L, Guyon-Debast A, Kermarrec M-P, Chauvin L, Chauvin J-E, Gallois J-L, Mazier M, Nogué F (2020b) Expanding the CRISPR toolbox in P. patens using SpCas9-NG variant and application for gene and base editing in Solanaceae Crops. Int J Mol Sci 21. https://doi.org/10.3390/ijms21031024
Voytas DF (2013) Plant genome engineering with sequence-specific nucleases. Annu Rev Plant Biol 64:327–350. https://doi.org/10.1146/annurev-arplant-042811-105552
Walton RT, Christie KA, Whittaker MN, Kleinstiver BP (2020) Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368:290–296. https://doi.org/10.1126/science.aba8853
Wang M, Lu Y, Botella JR, Mao Y, Hua K, Zhu J-K (2017a) Gene targeting by homology-directed repair in rice using a Geminivirus-based CRISPR/Cas9 system. Mol Plant 10:1007–1010. https://doi.org/10.1016/j.molp.2017.03.002
Wang M, Mao Y, Lu Y, Tao X, Zhu J-K (2017b) Multiplex gene editing in rice using the CRISPR-Cpf1 system. Mol Plant 10:1011–1013. https://doi.org/10.1016/j.molp.2017.03.001
Wang J, Meng X, Hu X, Sun T, Li J, Wang K, Yu H (2019) xCas9 expands the scope of genome editing with reduced efficiency in rice. Plant Biotechnol J 17:709–711. https://doi.org/10.1111/pbi.13053
Wang X, Ye L, Lyu M, Ursache R, Löytynoja A, Mähönen AP (2020a) An inducible genome editing system for plants. Nat Plants 6:766–772. https://doi.org/10.1038/s41477-020-0695-2
Wang S, Zong Y, Lin Q, Zhang H, Chai Z, Zhang D, Chen K, Qiu J-L, Gao C (2020b) Precise, predictable multi-nucleotide deletions in rice and wheat using APOBEC-Cas9. Nat Biotechnol 38:1460–1465. https://doi.org/10.1038/s41587-020-0566-4
Wang M, Xu Z, Gosavi G, Ren B, Cao Y, Kuang Y, Zhou C, Spetz C, Yan F, Zhou X, Zhou H (2020c) Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol J 18:1645–1647. https://doi.org/10.1111/pbi.13330
Wang Q, Alariqi M, Wang F, Li B, Ding X, Rui H, Li Y, Xu Z, Qin L, Sun L, Li J, Zou J, Lindsey K, Zhang X, Jin S (2020d) The application of a heat-inducible CRISPR/Cas12b (C2c1) genome editing system in tetraploid cotton (G. hirsutum) plants. Plant Biotechnol J. https://doi.org/10.1111/pbi.13417
Wolter F, Puchta H (2018) The CRISPR/Cas revolution reaches the RNA world: Cas13, a new Swiss Army knife for plant biologists. Plant J 94:767–775. https://doi.org/10.1111/tpj.13899
Wolter F, Puchta H (2019) In planta gene targeting can be enhanced by the use of CRISPR/Cas12a. Plant J 100:1083–1094. https://doi.org/10.1111/tpj.14488
Wolter F, Klemm J, Puchta H (2018) Efficient in planta gene targeting in Arabidopsis using egg cell-specific expression of the Cas9 nuclease of Staphylococcus aureus. Plant J 94:735–746. https://doi.org/10.1111/tpj.13893
Wu F, Qiao X, Zhao Y, Zhang Z, Gao Y, Shi L, Du H, Wang L, Zhang Y-J, Zhang Y, Liu L, Wang Q, Kong D (2020) Targeted mutagenesis in Arabidopsis thaliana using CRISPR-Cas12b/C2c1. J Integr Plant Biol 62:1653–1658. https://doi.org/10.1111/jipb.12944
Xu R, Li J, Liu X, Shan T, Qin R, Wei P (2020) Development of plant prime-editing systems for precise genome editing. Plant Commun 1:100043. https://doi.org/10.1016/j.xplc.2020.100043
Xu Z, Kuang Y, Ren B, Yan D, Yan F, Spetz C, Sun W, Wang G, Zhou X, Zhou H (2021) SpRY greatly expands the genome editing scope in rice with highly flexible PAM recognition. Genome Biol 22:6. https://doi.org/10.1186/s13059-020-02231-9
Yan F, Kuang Y, Ren B, Wang J, Zhang D, Lin H, Yang B, Zhou X, Zhou H (2018) Highly efficient A·T to G·C base editing by Cas9n-Guided tRNA Adenosine deaminase in rice. Mol Plant 11:631–634. https://doi.org/10.1016/j.molp.2018.02.008
Yu Y, Wang X, Sun H, Liang Q, Wang W, Zhang C, Bian X, Cao Q, Li Q, Xie Y, Ma D, Li Z, Sun J (2020) Improving CRISPR-Cas-mediated RNA targeting and gene editing using SPLCV replicon-based expression vectors in Nicotiana benthamiana. Plant Biotechnol J 18:1993–1995. https://doi.org/10.1111/pbi.13384
Zeng D, Li X, Huang J, Li Y, Cai S, Yu W, Li Y, Huang Y, Xie X, Gong Q, Tan J, Zheng Z, Guo M, Liu Y-G, Zhu Q (2020) Engineered Cas9 variant tools expand targeting scope of genome and base editing in rice. Plant Biotechnol J 18:1348–1350. https://doi.org/10.1111/pbi.13293
Zetsche B, Gootenberg JS, Abudayyeh OO, Slaymaker IM, Makarova KS, Essletzbichler P, Volz SE, Joung J, van der Oost J, Regev A, Koonin EV, Zhang F (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell 163:759–771. https://doi.org/10.1016/j.cell.2015.09.038
Zhang D, Zhang H, Li T, Chen K, Qiu J-L, Gao C (2017) Perfectly matched 20-nucleotide guide RNA sequences enable robust genome editing using high-fidelity SpCas9 nucleases. Genome Biol 18:191. https://doi.org/10.1186/s13059-017-1325-9
Zhang Q, Xing H-L, Wang Z-P, Zhang H-Y, Yang F, Wang X-C, Chen Q-J (2018) Potential high-frequency off-target mutagenesis induced by CRISPR/Cas9 in Arabidopsis and its prevention. Plant Mol Biol 96:445–456. https://doi.org/10.1007/s11103-018-0709-x
Zhang Y, Malzahn AA, Sretenovic S, Qi Y (2019) The emerging and uncultivated potential of CRISPR technology in plant science. Nat Plants 5:778–794. https://doi.org/10.1038/s41477-019-0461-5
Zhang X, Chen L, Zhu B, Wang L, Chen C, Hong M, Huang Y, Li H, Han H, Cai B, Yu W, Yin S, Yang L, Yang Z, Liu M, Zhang Y, Mao Z, Wu Y, Liu M, Li D (2020) Increasing the efficiency and targeting range of cytidine base editors through fusion of a single-stranded DNA-binding protein domain. Nat Cell Biol 22:740–750. https://doi.org/10.1038/s41556-020-0518-8
Zhong Z, Zhang Y, You Q, Tang X, Ren Q, Liu S, Yang L, Wang Y, Liu X, Liu B, Zhang T, Zheng X, Le Y, Zhang Y, Qi Y (2018) Plant genome editing using FnCpf1 and LbCpf1 Nucleases at redefined and altered PAM sites. Mol Plant 11:999–1002. https://doi.org/10.1016/j.molp.2018.03.008
Zhong Z, Sretenovic S, Ren Q, Yang L, Bao Y, Qi C, Yuan M, He Y, Liu S, Liu X, Wang J, Huang L, Wang Y, Baby D, Wang D, Zhang T, Qi Y, Zhang Y (2019) Improving plant genome editing with high-fidelity xCas9 and non-canonical PAM-targeting Cas9-NG. Mol Plant 12:1027–1036. https://doi.org/10.1016/j.molp.2019.03.011
Zhu H, Li C, Gao C (2020) Applications of CRISPR-Cas in agriculture and plant biotechnology. Nat Rev Mol Cell Biol 21:661–677. https://doi.org/10.1038/s41580-020-00288-9
Acknowledgments
We apologize to all colleagues in this rapidly growing field of genome engineering, whose work, due to space limitations, we were not able to cite. This work was supported by the EU Horizon 2020 Project 760331-2, ‘Newcotiana’. We want to thank Michelle Rönspies for critical reading of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.. This work was funded by the EU Horizon 2020 Project 760331–2 ‘Newcotiana’.
Author information
Authors and Affiliations
Contributions
All authors wrote, read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Consent for publication
authors consent to publish this review in Transgenic Research.
Code availability
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Huang, TK., Puchta, H. Novel CRISPR/Cas applications in plants: from prime editing to chromosome engineering. Transgenic Res 30, 529–549 (2021). https://doi.org/10.1007/s11248-021-00238-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-021-00238-x