Abstract
Genetic manipulation of carotenoid biosynthesis in higher plants has been the objective of a number of biotechnology programs, e.g. the Golden Rice Program. However, tomato (Solanum lycopersicum L.), which naturally accumulates lycopene in fruits, has attracted the attention of many groups who have manipulated it to increase or diversify carotenoid accumulation. One of the most significant achievements was “HighCaro (HC),” a transgenic tomato plant constitutively expressing the tomato lycopene beta-cyclase (tLcy-b), that produces orange fruits due to the complete conversion of lycopene to β-carotene. In this article we report the results of a field trial conducted in Metaponto (Italy) on HC and on two control genotypes to evaluate the stability of the transgenic trait and their yield performances. Transcriptional regulation of eight genes involved in carotenogenesis was assayed by quantitative real-time PCR (qRT-PCR) analysis on fruits collected at four distinct development stages. Statistical analysis results demonstrated that in field conditions the transgene maintained its ability to induce the conversion of lycopene to β-carotene. Moreover, agronomic performances and fruit quality in the transgenic line were not impaired by this metabolic disturbance. Results of qRT-PCR analysis suggested that transcription of PSY-1, PDS and ZDS genes were developmentally regulated in both genotypes. Unexpectedly, Lcy-b expression in transgenic fruits was also developmentally regulated, despite the fact that the gene was driven by a constitutive promoter. Our data provide evidence that in photosynthetic cells a strict and aspecific mechanism controls the level of transcripts until the onset of chromoplasts differentiation, at which point a gene-specific control on transcription takes place.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
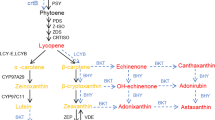
Carotenoids are an important class of natural pigments, which are mainly found in plants, yeast, algae and bacteria (Britton 1995). The basic chemical structure consists of eight isoprenoid units joined end to end to form a C40 hydrocarbon skeleton that included a system of conjugated double bonds (the chromophore) and linear or cyclic end groups. These characteristics account for the physical, chemical and biological properties. Carotenoids are divided into two groups: carotenes, containing only carbon and hydrogen atoms and xanthophylls, which contain oxygen atoms in various chemical forms (keto, hydroxyl, epoxy, etc.). Plant carotenoids are accumulated in plastids and play a fundamental role in photosynthesis as accessory pigments and in the xantophyll cycle, where they prevent photo-oxidative damage. They accumulate in chromoplasts in flowers and fruits, making these organs attractive to insects and other animals, thus facilitating pollination and seed dispersal. More than 700 different carotenoids and geometrical isomers have been described (EFSA FEEDAP panel 2005). In addition to their colouring properties, 50 carotenoids are also known to have provitamin A activity and are fundamental for the animals, including humans, which cannot synthesise this vitamin. β-Carotene, in particular, has the greatest provitamin A activity. Lycopene has been shown to posses a strong antioxidant action while lutein and zeaxanthin are deposited in the eyes and protect retinal cells of the macula against “phototoxic” damage caused by short-wavelength high-energy light radiation (Finley 2005; Omoni and Aluko 2005). Carotenoid biosynthesis in higher plants has been extensively studied both in model species (tobacco and Arabidopsis) and crop plants (tomato, pepper, potato) but several aspects of gene expression control, mainly post-transcriptional and post-translational, are still poorly understood (Hirschberg 2001; Fraser and Bramley 2004). Ever since the structure of β-carotene was elucidated, carotenoids have attracted the attention of industry and much effort has been put into chemical synthesis. Nevertheless, to date few commercially important carotenoids are synthesised chemically. Astaxanthin and β-carotene are included in this short list but the commercial products that contain them as active ingredients are mainly derived from microorganism cultures and in particular the alga Dunaliella spp. for β-carotene and the yeast Phaffia rhodozyma and the alga Haematococcus pluvialis for the astaxanthin. Zeaxanthin is currently obtained by extraction from corn grain and corn gluten meal. β-Carotene is used as a human dietary supplement for its provitamin A activity and for its antioxidant properties. Moreover it is used as a natural food colouring for a range of food and beverages such as cheese, fruit juices, margarine, etc. For the same reasons β-carotene is also used in pet foods mainly for dogs, cats, fish and birds (Dufossé et al. 2005). Zeaxanthin and astaxanthin are used as feed additives to add colour to the final products. The poultry industry uses zeaxanthin in feed to impart colour in yolk, skin, shanks and beaks. Astaxanthin is widely used in the salmonid industry (salmona and trout) as feed supplement because it is deposited in the flesh, giving the characteristic red-pink colour which is perceived by the consumer as one of the most important quality parameters. Moreover, astaxanthin has several other beneficial effects on farmed fishes, including antioxidant and provitamin A activity, and positive effects on reproduction, larval development, etc. Carotenoids today are obtained mainly from microorganism cultures and chemical synthesis. However, there are a number of specific drawbacks inherent to these processes that justify the development of more efficient, economical and specific plant-based systems. Several examples of successful experiments are now available that demonstrate that transgenic plants may be used to accumulate carotenoids in specific tissues or organs (Mann et al. 2000; Dharmapuri et al. 2002; Ralley et al. 2004; Davuluri et al. 2005). One of the most interesting achievements is the HighCaro (HC) tomato plant (D’Ambrosio et al. 2004). This transgenic line carries the tomato lycopene betacyclase (tLcy-b) cDNA driven by the CaMV 35S promoter and produces orange fruits as a result of the complete conversion of lycopene to β-carotene. HC has been extensively evaluated in greenhouse conditions and the transgene has been shown to be stably inherited.
In this article, we report the results of an openfield trial conducted in Metaponto (Basilicata Region, Italy) in 2004 to evaluate the stability of the transgenic trait and yield performances of the T6 homozygous HC line in comparison with two control lines: the isogenic Red Setter (RS) and the commercial hybrid Perfect Peel (HP). Transcription analysis of eight genes involved in the carotenoid biosynthesis in tomato was also conducted using replicated samples of HC and RS fruits at four stages of development.
Materials and methods
Plant materials, experimental design and data analysis
The trial was performed in Metaponto (40°23′32.16″N, 16°46′46.44″E) from April to August 2004 (Fig. 1) and complied in accordance with laws ruling the deliberate release in open fields of a genetically modified organism in the European Union. Three tomato (Solanum lycopersicum L.) genotypes were compared: the transgenic HC line (T6 generation), the cv RS, from which the HC was derived, and the commercial hybrid Perfect Peel (Seminis Vegetable Seeds Italia S.r.l.). The transgenic T6 generation was obtained by repeated cycles of self-pollination starting from the transgenic plant transformed with the tLyc gene via Agrobacterium tumefaciens (D’Ambrosio et al. 2004).
Open-field evaluation of HighCaro, Red Setter and Perfect Peel tomato lines conducted during Spring-Summer of 2004 in Metaponto (Southern Italy). (a) The experimental field with the surrounding fence and the label indicating the type of experimentation. (b) Biological containment made by four rows of maize and two rows of tomato plants surrounding the experimental plots. (c) Each plot was made by 90 plants distributed among three beds consisting of two rows of 15 plants each
A randomised complete block design (RCBD) was used with 12 replicates.
Each plot was 6 m × 5 m (L × W) with three beds consisting of two rows of 15 plants. The two rows were spaced 0.5 m apart while plants on the same row were planted 0.4 m apart. Each plot was 30 m2 and contained 90 plants at a density of 3 plants per m2. Biological and physical containment was obtained with four rows of maize and two rows of tomato plants surrounding the experimental plots. The trial was conducted following standard agronomical procedures. A single harvest was carried out at the end of the growing season. The number of plants in each of the 36 plots was recorded in order to express yield components on a plant base. Fruits were collected and subdivided into three groups to derive yield components: marketable fruits, immature fruits and non-marketable fruits. Overall agronomic performances were estimated on data from all 36 plots while chemical determinations were performed on a suitable amount of fruits sampled from the marketable yield of 24 plots. The 24 plots were chosen in such a way as to constitute a RCBD with eight replicates. Analysis of variance (ANOVA) was used to estimate the effects of the genotype factor on all measured traits while the Bonferroni t-test was used to verify the statistical significance of recorded differences. All statistical analyses were conducted using specific procedures of the STAT module of the SAS program (SAS® System for Windows ver. 9). Fruits not used for analyses were sterilised before disposal. Plants or parts of them were removed from the soil, left on the soil to dry and later burned on site. The test area was marked to monitor re-emergence of volunteer tomato plants over the following seasons.
Fruit quality and carotenoid determinations
Analysis of dry weight (DW), titratable acidity (acidity), soluble solids and conductivity were performed on fruit homogenate obtained by blending the fruits in a food processor. To measure DW, a sub-sample of the homogenate was weighed and dried in an oven set at 70°C until the weight was constant. Values were expressed as grams per 100 g of fresh weight (FW). The other analytical measurements were carried out on the liquid portion obtained after centrifuging the homogenate. Acidity was determined by titration with a Metrohm 654 pH-meter (Metrohm Ltd, Switzerland) and values were expressed in mEq of citric acid per 100 g. Soluble solids were determined using the digital refractometer ATAGO DBX-55 (ATAGO CO. LTD, Japan), which provides values as Brix degrees (°B). Conductivity was measured with the Metrohm Conductometer 660 (Metrohm Ltd, Switzerland) and expressed as μS/ cm. Carotenoids were extracted from ripe tomato fruits and analysed by HPLC, as described previously (D’Ambrosio et al. 2004).
Quantitative real-time PCR analysis
Relative quantification of transcript steady-state levels for eight genes encoding enzymes involved in the metabolism of carotenoids in tomato plants, was performed on leaves and on fruits at four stages of development for HC and RS cultivars.
Fruits were collected at four development stages (mature green, breaker, pink and ripe) (Bedbrook et al. 1999) from four plots for each genotype in such a way as to fulfil the requirements for a RCBD. For each RNA sample, at least eight fruits were collected from the same plot and cut in two parts to inspect the pericarp colour (Fig. 2). A piece of pericarp from the four most uniform fruits was excised to form the bulk for total RNA extraction (López-Gómez and Gómez-Lim 1992). Leaves for total RNA extraction were collected from the same plots on plants prior to flowering. DNA contamination was removed from RNA extracts using the DNA-Free kit (Ambion, Austin, TX, USA). A PCR was carried out with a pair of primers specific for the 18S rRNA gene (forward: GCATTTGCCAAGGATGTTTT; reverse: TAGCAGGCTGAGGTCTCGTT) to test the efficiency of DNA removal.
Fruits of transgenic (HighCaro) and control (Red Setter) lines sampled at the mature green and the ripe stages for the extraction of RNA. (a) Fruits at mature green stage from the plots 25 (Red Setter) and 27 (HighCaro). Fruits of HighCaro (b) and Red Setter (c) lines at ripe stage sampled from the four replicates
Complementary DNA was synthesised from 1.5 μg of RNA using the ThermoScript™ RT-PCR System kit (Invitrogen) with random hexamer primers following the manufacturer’s instructions. Reactions were conducted in 96-well reaction plates of 25 μl volume containing 12.5 μl of the Platinum® Quantitative PCR SuperMix-UDG (Invitrogen), 300 nM forward primer, 300 nM reverse primer, 150 nM TaqMan probe. One microlitre of the cDNA sample (75 ng of RNA) was used for each reaction, except for Lcy-b, in which the amount was doubled.
Quantitative real-time PCR analysis (qRT-PCR) was performed using the iCycler iQ™ Real-Time PCR Detection System (Bio-Rad Laboratories Inc., USA). Thermal cycling condition were 95°C for 3 min for activation of DNA polymerase and 40 cycles of 95°C for 15 s and 60°C for 1 min.
Estimates of transcript content were derived using the standard curve method (User Bulletin #2, ABI PRISM 7700 Sequence Detection System, Relative quantitation of gene expression, 2001, AB Applied Biosystems) performing reactions in separate tubes. Standard curves were prepared for both the target transcripts and the endogenous reference 18S rRNA gene by assembling a set of reactions using threefold serial dilutions of a fruit cDNA stock sample. Each PCR reaction was performed in triplicate, both for the construction of the standard curve and for sample quantitation. Two plates were run for each gene and both contained a plate-specific standard curve to account for plate variation estimates. Gene starting quantities (SQs) for each sample were estimated using regression parameter estimates of the standard curve. The values were then divided by the corresponding 18S rRNA gene SQs in order to normalise results for differences in the amount of total RNA content among samples. The genes selected for the analysis were the following: fruit-specific phytoene synthase (PSY-1); leaf-specific phytoene synthase (PSY-2), phytoene desaturase (PDS), zetacarotene desaturase (ZDS), lycopene beta-cyclase (Lcy-b), carotene beta-hydroxylase 1 (CrtR-b1), zeaxanthin epoxydase (ZEP), 9-cis-epoxycarotenoid dioxygenase 1 (NCED1), neoxanthin synthase (nxs) also known as chromoplast-specific lycopene cyclase (Cyc-b) and the 18S rRNA gene, which was used as internal standard. Primer pairs and TaqMan® probes were designed with Primer Express software (PE-Applied Biosystems). Probes were labelled at the 5′ end with carboxyfluorescein (FAM) and at the 3′ end with carboxytetramethylrhodamine (TAMRA). Table 1 in the Supplementary material reports sequences of primers and probes for the gene assayed. Statistical significances of the difference between the means for the two genotypes for each gene and stage combination were evaluated using Student’s t-test and the null hypothesis was rejected at the 0.05 level.
Results
An open-field trial was conducted (under the authorization DEC/DPN/651 of 26th April 2004 issued by the “Ministero dell’ambiente e della tutela del territorio” of Italy) on the transgenic HC tomato line and control varieties in Metaponto (Basilicata Region, Italy) in 2004 with the aim of evaluating the stability of the transgenic trait and the field performances of the T6 homozygous HC line in comparison to the RS isogenic control variety and the Perfect Peel (HP), a highly productive commercial hybrid (Seminis Vegetable Seeds Italia S.r.l.).
Yield components
A single harvest was carried out at the end of August and from each of the 36 plots belonging to 12 blocks, the number of plants, the weight of marketable, immature and unmarketable (rotted or infested) fruits were recorded and four yield components were calculated and expressed as grams per plant. Statistical analysis (Table 1) indicated that the genotype factor had a significant effect on total yield (TY, the sum of marketable, immature and unmarketable fruits), marketable (MY) and unmarketable yield (UMY), but not on immature fruits yield (IFY). The Perfect Peel hybrid displayed, as expected, the best yield performances producing an average 949.8 g of fruit per plant, followed by RS with 725.9 g and HC with 431.2 g. All three genotype mean differences were statistically significant for this trait. The MY mean for HP (734.5 g/plant) was statistically different from that of RS (450.4 g/plant) and that of HC (288.5 g/plant) but the two isogenic line means did not differ significantly between themselves. Conversely, the difference between the means of UMY for HC and RS (respectively, 78.1 and 178.1 g/plant) was statistically significant with the transgenic line having the lower and most appreciable value. No difference among the three genotypes was shown for the last yield component, the weight of immature fruit per plant. We also analysed MY, UMY and IFY expressed as a percentage of TY. Again HP performed better than RS and HC. The Perfect Peel hybrid displayed the highest value of MY (77%) and the lowest value both of UMY (9%) and IFY (13%). The two isogenic lines tended to have similar values for MY (62 and 67%) and for UMY (13 and 15%), while a slightly higher proportion of immature fruits was recorded for the control RS variety (24%) compared to the transgenic HC line (18%).
Carotenoid contents and other fruit quality parameters
One of the main objectives of this experiment was to evaluate the stability of the transgene expression in open-field conditions. The HC line was obtained by transforming the tomato cv RS with a gene cassette containing the tomato lycopene beta-cyclase cDNA (Lcy-b) driven by CaMV 35S promoter (D’Ambrosio et al. 2004). Fruits of this line were shown to convert virtually all lycopene to β-carotene and appeared orange instead of red. This transgene has been shown to be stably inherited with a complete penetrance from the starting T0 mother plant up to generation T5 in greenhouse conditions.
Carotenoid contents of ripe fruits estimated through HPLC analysis (Table 2) showed that HC fruits also converted almost all lycopene to β-carotene in open-field conditions. ANOVA results showed that genotype treatment effects were statistically significant for all the carotenoid content variables. Mean separation results showed that the HC mean was statistically different from the means of RS and HP for all tested variables. HC fruits contained a very high content of β-carotene and a very low content of lycopene and lutein compared to RS and HP fruits, which did not differ between themselves for any of the tested variables. HC fruits contained on average 76.6 μg of β-carotene, 0.5 μg of lycopene and 2.3 μg of lutein per g of FW. RS and HP fruits contain, respectively, 6.6 and 5.7 μg of β-carotene, 102.9 and 108.9 μg of lycopene and 10.9 and 11.4 μg of lutein per g of FW. Total carotenoid content was lower in HC (84.0 μg) than in RS and HP fruits (125.4 and 131.7 μg).
HC and HP fruits resulted quite similarly in terms of DW, titratable acidity and conductivity of homogenate, while RS showed slightly lower values for these parameters. Homogenate DW was 5.08% for RS fruits and 5.58 and 5.64% for HC and HP fruits. Titratable acidity was 5.41 for RS, 6.12 for HC and 5.95 for HP fruits. The soluble solids value of HC fruit homogenate was the highest (4.36°B), that of RS was the lowest (3.96°B) while that of HP was intermediate between the two (4.13°B). For this trait, the means of the three genotypes differed significantly, while for the other parameters the RS differed significantly from HP and HC, which did not differ significantly between themselves.
Transcriptional analysis of carotenoid biosynthesis in developing tomato fruits
Transcript quantification of eight genes (PSY-1, PDS, ZDS, Lcy-b, CrtR-b1, ZEP, VDE1 and Cyc-b/nxs), in the pericarp of RS and HC fruits was performed at four stages of maturation (mature green, breaker, pink and ripe), using Real-Time PCR analysis with gene-specific fluorescent probes. Leaf RNA transcript contents for the same genes, with the exception of PSY-1, which was replaced from the leaf-specific gene (PSY-2), were also estimated from RNA extracted from leaves collected before flowering. Gene transcript estimates were expressed as a percentage of the endogenous reference 18S rRNA gene transcript estimate for each sample (normalisation), thus producing a unitless number that could be used to make comparisons among samples and genes (Table 3).
Transcription patterns of the eight genes involved in carotenoid biosynthesis during maturation of RS and HC fruits are shown in Fig. 3aFig. 3a, Fig. 3b. Estimated transcript levels in leaf RNA are presented in Fig. 4.
The PSY-1 gene was shown to have a distinct expression pattern, in agreement with prior reports based on different quantification methods (Ronen et al. 1999; Giuliano et al. 1993; Pecker et al. 1996; Alba et al. 2005). Its transcript was almost absent at the mature green stage, as at this stage fruit tissues may be considered equivalent to leaf tissues, where only PSY-2 is expressed.
PSY-1 transcription increased dramatically between the breaker and pink stages, where it reached the highest values for both the genotypes (1.58 and 1.22 for HC and RS, respectively). At the ripe stage a slight reduction was observed for both genotypes. The difference between the genotype’s means at the four stages were not significant. The PDS and ZDS patterns in RS were quite similar, showing a gradual increase from mature green until the pink stage and a slight reduction at the ripe stage. HC patterns for the same genes were instead different. The transcripts increased until the ripe stage. The only statistically significant difference between the two genotypes was for ZDS at the ripe stage, with the HC line showing greater transcript values. As expected, Lcy-b patterns were the most different between the two genotypes. The over-expression of the Lcy-b transgene in the HC genotype was quite evident, since its transcript was present in much greater levels at all stages in the transgenic than in control RS RNA. At all stages, the means of the two genotypes were significantly different. However, the transcription pattern of Lcy-b in the HC genotype was somewhat unexpected. The transcript level was relatively low at the mature green stage and increased up to the ripe stage, where it seemed to reach a plateau. On the other hand, the Lcy-b transcription pattern in RS showed a gradual reduction between the mature green, the breaker and the pink stage followed by a small increase at the ripe stage.
Carotene b-hydroxylase 1 (CrtR-b1) transcript patterns were quite similar in the two genotypes. Gene transcription increased gradually between mature green and the ripe stage. However, the transcription in HC fruits was significantly greater that in RS fruits at the ripe stage. The ZEP gene was also shown to have a similar pattern in the two genotypes, the only significant difference occurring at the mature green stage. However, this was the only gene for which transcript levels were lower in HC fruits than in RS fruits at all the stages. NCED1 and Cyc-b/nxs transcription was very low compared to the other genes in both genotypes during fruit maturation and no significant difference was visible between the two genotypes. However, while NCED1 seemed to gradually decrease between the mature green and ripe stage, Cyc-b/nxs transcription gradually increased during fruit maturation for both the genotypes. Comparison of transcript level estimates for the eight genes in leaf RNA in the two genotypes (Fig. 4) showed that the most abundant transcript was PSY2, followed by ZEP and finally by CrtR-b1, ZDS, PDS, NCED1 and Cycb/nxs. Lcy-b transcription was low in HC leaves and very low in the RS leaves. In order to compare gene transcription levels in leaves with levels in green fruit tissues, we divided the leaf transcription level by the transcription level in fruits at the mature green stage. This value was calculated for all genes except phytoene synthase. This analysis showed that the contents of NCED1 and Cyc-b/nxs were between two- and eightfold lower in leaves than in fruits at the mature green stage. Conversely, ZDS transcription was about five- and fourfold greater in leaves than in mature green fruits, respectively, for HC and RS. The most interesting results of this analysis concerned the ZEP gene. ZEP transcription was tenfold greater in leaves than in mature green fruits in the HC genotype. On the other hand, the RS leaf value was only 2.7 times greater than the fruit value, probably because at the mature green stage the ZEP transcript was significantly greater in RS fruits than in HC fruits, while in leaf RNA the transcription means of the two genotypes were not statistically different.
Real-time PCR analysis of transcript contents of the eight carotenoid genes in transgenic fruits (HighCaro) and control (Red Setter) fruits at four developmental stages. (a) PSY-1 phytoene synthase, PDS phytoene desaturase, ZDS zeta-carotene desaturase, lcy-b lycopene beta-cyclase. (b) CrtR-b1 beta-carotene hydroxylase 1, ZEP zeaxanthin epoxidase, NCED1 9-cis-epoxycarotenoid dioxygenase, nxs/cyc-b neoxanthin synthase/chromoplast-specific lycopene cyclase b. The error bar is the sample mean standard error (N = 4). An asterisk indicates that the difference between the means of the two genotypes at that stage was statistically significant (Student’s t-test; α = 0.05)
Discussion
An open-field trial was conducted in 2004 in Metaponto (Southern Italy) on the transgenic tomato line HC, with the aim of evaluating its agronomical performance and the stability and penetrance of the tLcy-b transgene. A suitable experimental design was employed to make reliable comparisons between the transgenic line, its isogenic RS variety and the commercial hybrid Perfect Peel. Also, fruits were collected from replicated plots of HC and RS genotypes at four different stages of development in order to study the transcriptional regulation of carotenoid accumulation in tomato fruits.
The field trials demonstrated that even in open-field conditions, the HC fruits contained large amounts of β-carotene as a result of the lycopene beta-cyclase’s cyclisation activity. Agronomical performances of the transgenic line were comparable to those of the control RS line though both performed worse than the tester hybrid HP. Although the HC line showed a significantly lower value of total fruit weight per plant (TY), the two lines did not differ in terms of marketable fruits (MY) production, while for RS a significantly higher value of unmarketable fruit weight per plant was recorded.
As for carotenoid contents, transgenic fruits contained significantly less total carotenoids and lutein than RS and Perfect Peel hybrid fruits. These results are not surprising, as it had been already reported (D’Ambrosio et al. 2004) that growing conditions greatly influence performance of the HC line. In particular, transgenic fruits grown at high temperatures in a greenhouse were shown to contained reduced amounts of carotenoids. As for lutein content, the reduced amount in transgenic compared to control fruits can have a simple biochemical explanation. If we suppose that the high content of Lcy-b transcript is in all likelihood followed by a high content of the encoded enzyme, we conclude that lycopene is preferentially β-cyclised rather than ε-cyclised and therefore the synthesis of lutein is ill-favoured compared to the synthesis of β-carotene derived xanthophylls.
Statistical analysis of homogenate parameters (DW, soluble solids, titratable acidity and conductivity) showed that transgenic fruits have a greater content of DW, soluble solids and titratable acidity.
Altogether, the results of fruit quality characterisation in open-field conditions of the HC line in comparison to isogenic RS and the hybrid Perfect Peel, demonstrated that the transgene maintains its ability to induce a substantial conversion of lycopene to β-carotene in developing fruits. Thus, the effects of the transgene seemed to be confined to what was expected, that is, a modification of the fruit carotenoid contents and of correlated indices such as soluble solids.
Transcriptional control of carotenoid biosynthesis in developing tomato fruits
Comparative analysis of transcription patterns for eight genes involved in carotenoid biosynthesis in tomato fruits gives a new insight into the coordinated control of genes involved in this process. Results of qRT-PCR suggested that the transcription of PSY-1, PDS and ZDS genes, encoding enzymes involved in the biosynthesis of lycopene, was developmentally regulated (Fig. 5). All genes showed a similar pattern: transcripts started to increase at the breaker stage, reached a peak at the pink stage and then decreased slightly until the ripe stage. PSY-1 was the gene with the highest level of transcription, followed by PDS and then by ZDS. At the pink stage, PSY-1 transcription was almost 100 times higher than at the mature green stage, while PDS and ZDS transcription increased fivefold between these two stages (Table 3). Our results are highly consistent with reports by other authors (Ronen et al. 1999; Giuliano et al. 1993; Pecker et al. 1992, Pecker et al. 1996; Corona et al. 1996; Alba et al. 2005). qRTPCR of HC fruits showed that PSY-1, PDS and ZDS transcription increases at the breaker stage, but while PSY-1 and PDS peak at the pink stage, ZDS transcription continued to increase until the ripe stage.
Comparison of the lycopene beta-cyclase gene (lcy-b) transcription pattern in the two genotypes allowed us to draw some new and interesting conclusions. It had been suggested that the accumulation of lycopene in tomato occurring after the breaker stage is the result of the down-regulation of Lcy-b and Lcy-e transcription (Pecker et al. 1996; Ronen et al. 1999; van den Berg et al. 2000; Alba et al. 2005). Our data are consistent with this interpretation, since further reduction of the already low Lcy-b transcription was observed between the breaker and the pink stage in RS fruits. However, analysis of the Lcy-b expression pattern in developing HC fruits complicated the scenario as the transgene was also shown to be developmentally regulated. At the mature green stage, HC fruit RNA contained the lowest amount of Lcy-b transcript, which increased gradually as the fruit developed. This transcription pattern was completely unexpected, considering the constitutive CaMV 35S promoter. Transcription was expected to be constant throughout fruit development. A plausible explanation could be the presence of a post-transcriptional mechanism in chloroplast-containing cells in tomato plants. While in control fruits all available data indicated that Lcy-b expression was transcriptionally controlled from breaker stage onward, HC fruit data seemed to show aspecific post-transcriptional control on mRNA levels, as demonstrated by the fact that Lcy-b transcript was kept low until the breaker stage.
Moreover, the steady increase of Lcy-b transcription during maturation in HC fruits was paralleled by the increase of other transcripts, that is PDS, ZDS and CrtR-b1, demonstrating a transcription induction of endogenous carotenoid biosynthetic genes, similar to that observed in tomato fruits over-expressing a bacterial carotene desaturase gene CRTI (Römer et al. 2000).
As a whole, our data provide evidence that in photosynthetic cells the transcription of genes is under a strict and aspecific control mechanism in such a way that mRNAs with unusually high levels are partially degraded in order to prevent potential impairment of the photosynthetic apparatuses. When the radical changes from chloroplast to chromoplast take place, greater RNA levels are tolerated in order to sustain carotenogenesis, but specific transcriptional control on Lcy-b RNA mediated by ethylene (Alba et al. 2005) must be activated to permit lycopene accumulation. In this regard, it is notable that the Lcy-b transgene transcript content estimated in HC leaf RNA is only 41% of that reached in the fruit of the same genotype at the ripe stage.
An interesting point of discussion concerns the expression pattern of the chromoplast-specific lycopene cyclase (Cyc-b) gene, also characterised as a nxs gene (Ronen et al. 2000; Bouvier et al 2000). It has been demonstrated that this gene encodes a bi-functional enzyme capable of converting both lycopene to β-carotene and violaxanthin to neoxanthin. Our data showed identical Cyc-b/nxs expression patterns in RS and HC fruits. In particular, gene transcription increased slowly throughout fruit development. These results are not in contrast with Ronen et al. (Ronen et al. 2000), who observed that after the breaker stage a new pool of β-carotene is synthesised by the Cyc-b encoded cyclase in wild-type tomato. Conversely, the high β-carotene content in HC transgenic fruits was almost entirely a result of the lycopene beta-cyclase encoded by the Lcy-b transgene.
Final considerations focus on the transcription pattern of the ZEP gene, encoding the enzyme ZEP. ZEP transcription patterns were similar in the two genotypes and were characterised by a peak at the breaker stage and a minimum at the pink stage. Another peculiarity of this pattern, compared to those of the other genes, was that only in this case was relative transcription higher in RS fruits than in transgenic fruits during all stages, particularly the mature green stage.
In summary, this study carried out on replicated experimental fruit samples both from control and transgenic lines grown in open-field conditions, allowed us to demonstrate that in these conditions the lycopene cyclase transgene maintains its ability to induce a substantial conversion of lycopene to β-carotene in developing fruits. Moreover, the analysis in RS and transgenic HC developing fruits of the transcription patterns of eight genes involved in the biosynthesis of carotenoids seemed to indicate that in chloroplast-containing cells the contents of exogenous and endogenous RNAs are tightly controlled and that this control is released with the onset of chromoplasts differentiation.
References
Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17:2954–2965
Bedbrook RJ, Dunsmuir P, Howie JW, Joe KL, Lee YK (1999) Delayed ripening tomato plants with T-DNA bearing a truncated ACC2 synthase gene. US Patent 5,952,546
van den Berg H, Faulks R, Fernando Granado H, Hirschberg J, Olmedilla B, Sandmann G, Southon S, Stahl W (2000) The potential for the improvement of carotenoid levels in foods and the likely systemic effects. J Sci Food Agric 80:880–912
Bouvier F, D’Harlingue A, Backhaus RA, Kumagai MH, Camara B (2000) Identification of neoxanthin synthase as a carotenoid cyclase paralog. Eur J Biochem 267:6346–6352
Britton G (1995) Structure and properties of carotenoids in relation to function. FASEB J 9:1551–1558
Corona V, Aracri B, Kosturkova G, Bartley GE, Pitto L, Giorgetti L, Scolnik PA, Giuliano G (1996) Regulation of a carotenoid biosynthesis gene promoter during plant development. Plant J 9:505–512
D’Ambrosio C, Giorio G, Marino I, Merendino A, Petrozza A, Salfi L, Stigliani AL, Cellini F (2004) Virtually complete conversion of lycopene into beta-carotene in fruits of tomato plants transformed with the tomato lycopene β-cyclase (tlcy-b) cDNA. Plant Sci 166:207–214
Davuluri GR, van Tuinen A, Fraser PD, Manfredonia A, Newman R, Burgess D, Brummell DA, King SR, Palys J, Uhlig J, Bramley PM, Pennings HM, Bowler C (2005) Fruit-specific RNAi-mediated suppression of DET1 enhances carotenoid and flavonoid content in tomatoes. Nat Biotechnol 23:890–895
Dharmapuri S, Rosati C, Pallara P, Aquilani R, Bouvier F, Camara B, Giuliano G (2002) Metabolic engineering of xanthophyll content in tomato fruits. FEBS Lett 22:30–34
Dufossé L, Galaup P, Yaron A, Arad SM, Blanc P, Chidambara Murthy KN, Ravishankar GA (2005) Microorganisms and microalgae as sources of pigments for food use: a scientific oddity or an industrial reality? Trends Food Sci Technol 16: 389–406
EFSA FEEDAP Panel (2005) Opinion of the Scientific Panel on Additives and Products or Substances used in Animal Feed on the request from the European Commission on the safety of use of colouring agents in animal nutrition PART I General Principles and Astaxanthin. EFSA J 291:1–40
Finley WJ (2005) Proposed criteria for assessing the efficacy of cancer reduction by plant foods enriched in carotenoids, glucosinolates, polyphenols and selenocompounds. Ann Bot 95:1075–1096
Fraser PD, Bramley PM (2004) The biosynthesis and nutritional uses of carotenoids. Prog Lipid Res 43:228–265
Giuliano G, Bartley GE, Scolnik P (1993) Regulation of carotenoid biosynthesis during tomato development. Plant Cell 5:379–387
Hirschberg J (2001) Carotenoid biosynthesis in flowering plants. Curr Opin Plant Biol 4:210–218
López-Gómez R, Gómez-Lim MA (1992) A method for extracting intact RNA from fruits rich in polysaccharides using ripe mango mesocarp. HortScience 27:440–442
Mann V, Harker M, Pecker I, Hirschberg J (2000) Metabolic engineering of astaxanthin production in tobacco flowers. Nat Biotechnol 18:888–892
Omoni OA, Aluko ER (2005) The anti-carcinogenic and anti-atherogenic effects of lycopene: a review. Trends Food Sci Technol 16:344–350
Pecker I, Chamovitz D, Linden H, Sandmann G, Hirschberg J (1992) A single polypeptide catalyzing the conversion of phytoene to ζ-carotene is transcriptionally regulated during tomato fruit ripening. Proc Natl Acad Sci USA 89:4962–4966
Pecker I, Gabbay R, Cunningham FX, Hirschberg J (1996) Cloning and characterization of the cDNA for lycopene β-cyclase from tomato reveals decrease in its expression during fruit ripening. Plant Mol Biol 30:807–819
Ralley L, Enfissi EM, Misawa N, Schuch W, Bramley PM, Fraser PD (2004) Metabolic engineering of ketocarotenoid formation in higher plants. Plant J 39:477–486
Römer S, Fraser PD, Kiano JW, Shipton CA, Misawa N, Schuch W, Bramley PM (2000) Elevation of the provitamin A content of transgenic tomato plants. Nat Biotechnol 18:666–669
Ronen G, Cohen M, Zamir D, Hirschberg J (1999) Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J␣17:341–351
Ronen G, Carmel-Goren L, Zamir D, Hirschberg J (2000) An alternative pathway to β-carotene formation in plant chromoplast discovered by map-based cloning of beta and old-gold color mutations in tomato. Proc Natl Acad Sci USA 97:11102–11107
Acknowledgements
The authors wish to thank all colleagues of Metapontum Agrobios who collaborated in the project and are indebted to Larisa Whitney for correcting the English. This project was conducted under the Metapontum Agrobios research program Safety Assessment in Food and Environment funded by the “Ministero dell’Istruzione, dell’Università e della Ricerca” of Italy.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License ( https://creativecommons.org/licenses/by-nc/2.0 ), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Giorio, G., Stigliani, A.L. & D’Ambrosio, C. Agronomic performance and transcriptional analysis of carotenoid biosynthesis in fruits of transgenic HighCaro and control tomato lines under field conditions. Transgenic Res 16, 15–28 (2007). https://doi.org/10.1007/s11248-006-9025-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11248-006-9025-3