Abstract
Three Ni(II) and Zn(II) complexes [Ni(L1)], [Ni(L2)], and [Zn(L3)(DMSO)] (L1 = 2,3-bis(2-hydroxybenzylideneimino)-2,3-butenedinitrile, L2 = 2,3-bis(2-hydroxy-3-methoxybenzylideneimino)-2,3-butenedinitrile, L3 = 2,3-bis(2-hydroxy-1-naphthylideneimino)-2,3-butenedinitrile) were obtained in DMSO by one-pot syntheses. The complexes were characterized by physicochemical and spectroscopic methods. Also, their solid-state structures were determined by single-crystal X-ray diffraction. The geometries of the Ni(II) and Zn(II) complexes were square planar and square pyramidal, respectively. The complexes were screened in vitro against a fungal species and eight species of bacteria, revealing their antimicrobial activity.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The synthesis and characterization of Schiff bases and their transition metal complexes has attracted considerable attention in recent decades [1,2,3]. Schiff base ligands with a variety of donor atoms including nitrogen, oxygen and sulfur can be coordinated with transition metal atoms. Depending on the type and oxidation state of the central metal atom and the ligand structure, they can be bidentate, tridentate, tetradentate or polydentate, and depending on the nature of the possible counter ion, transition metal Schiff’s base complexes show various coordination modes with varying degree of distortion [4, 5]. Among these, the tetradentate Schiff bases with N2O2 coordination sites belong to ligands investigated most often in coordination chemistry [6,7,8]. The transition metal complexes formed from tetradentate Schiff bases exhibit a wide range of biological properties [9,10,11,12]. 2,3-Diamino-cis-2-butenedinitrile (diaminomaleonitrile: DAMN) is an attractive precursors to nucleotides and is considered as an intermediate in the synthesis of a wide variety of heterocyclic compounds [13]. The condensation of DAMN with different aromatic aldehydes can form 1:1 and/or 1:2 Schiff base ligands [14]. In recent studies, we have reported the synthesis and characterization of complexes incorporating tridentate NNO-Schiff base ligands based on diaminomaleonitrile [15, 16]. In this project, our efforts were directed to the synthesis and characterization of nickel(II) and zinc(II) complexes with tetradentate N2O2-Schiff base ligands based on diaminomaleonitrile and various aldehydes such as 2-hydroxybenzaldehyde, 3-methoxy-2-hydroxybenzaldehyde and 2-hydroxy-1-naphthaldehyde.
Experimental
Materials and methods
All reagents for synthesis and analysis were purchased from Merck and used without further purification. Elemental analyses were recorded on a Thermo Finnigan Flash Elemental Analyzer 1112EA. Melting points were measured on an Electrothermal-9100 apparatus. The IR measurements were taken on a FTIR Tensor 27 infrared spectrophotometer as KBr disks in the range of 400–4000 cm−1. 1H NMR spectra were measured on a Bruker AVANCE BRX 250 MHz spectrometer using d6-DMSO as solvent for the complexes. The chemical shift values (δ) are given in ppm. The electronic absorption spectra in DMSO solution were recorded by a Cary 50 UV–Vis spectrophotometer. Bacteria were isolated from clinical samples or purchased from Merck Company.
Preparation of [Ni(L1)] and [Ni(L2)]
A solution of 2-hydroxybenzaldehyde (2.0 mmol, 0.21 ml) for H2L1 and 3-methoxy-2-hydroxybenzaldehyde (0.304 g, 2.0 mmol) for H2L2 and 2,3-diamino-cis-2-butenedinitrile (0.108 g, 1.0 mmol) in DMSO (5 ml) was heated for 30 min. Afterward, triethylamine (0.200 g, 2.0 mmol) and NiCl2·6H2O (0.238 g, 1.0 mmol) in DMSO (5 ml) were added dropwise and the resulting reaction mixture was refluxed for 4 h while stirring constantly. The clear green solution was allowed to stand several days at room temperature, providing blue single crystals suitable for X-ray crystallography (Scheme 1).
[Ni(L1)]: Yield: 0.242 g (65%); m.p. > 300 °C IR (KBr, cm−1): 2218 (C≡N), 1611 (C=N), 1576 (C=C)aliphatic, 1517 (C=C)aromatic, 1195 (C–O), 754 (Ni–N), 418 (Ni–O). 1H NMR (250 MHz, d6-DMSO, ppm): δ = 8.57 (s, 2H, –CH=N), 7.45–6.24 (m, 8H, ArH). UV–Vis spectra [λmax, nm (log ε, L mol−1 cm−1); DMSO solution]: 265 (4.46), 305 (4.30), 380 (4.25), 440 (4.08), 530 (4.09), 595 (4.08). Anal. Calcd. for C18H10N4NiO2: C, 57.9; H, 2.7; N, 15.0%. Found: C, 57.7; H, 2.6; N, 14.9%.
[Ni(L2)]: Yield 0.259 g (60%); m.p. > 300 °C. IR (KBr, cm−1): 2225 (C≡N), 1612 (C=N), 1580 (C=C)aliphatic, 1538 (C=C)aromatic, 1253 (C–O), 735 (Ni–N), 414 (Ni–O). 1H NMR (250 MHz, d6-DMSO, ppm): δ = 8.49 (s, 2H, –CH=N), 7.20–6.45 (m, 6H, ArH), 3.74 (s, 6H, –OCH3). UV–Vis spectra [λmax, nm (log ε, L mol−1 cm−1); DMSO solution]: 270 (4.29), 355 (4.16), 375 (4.13), 395 (4.14), 520 (3.97), 600 (4.13). Anal. Calcd. for C20H18N4NiO6: C, 51.2; H, 3.8; N, 11.9%. Found: C, 51.0; H, 3.7; N, 11.7%.
Preparation of [Zn(L3)(DMSO)]
This complex was synthesized similar to [Ni(L1)] and [Ni(L2)] complexes. To a solution of 2-hydroxy-1-naphthaldehyde (0.344 g, 2.0 mmol) in DMSO (10 ml), 2,3-diamino-cis-2-butenedinitrile (0.108 g, 1.0 mmol) was added and refluxed for 30 min. A deep brown solution was obtained. After adding triethylamine (0.200 g, 2.0 mmol) and Zn(CH3CO2)2·2H2O (0.219 g, 1.0 mmol) to the solution, the color of the solution changed to black. Dark red single crystals suitable for crystallography appeared after 2 weeks standing (Scheme 2). Yield 0.407 g (73%); m.p. > 300 °C. IR (KBr, cm−1): 3007 (C–H), 2209 (C≡N), 1601 (C=N), 1571 (C=C)aliphatic, 1536 (C=C)aromatic, 1267 (C–O), 1183 (S=O), 747 (Zn–N), 412 (Zn–O). 1H NMR (250 MHz, d6-DMSO, ppm): δ = 9.25 (s, 2H, –CH=N), 8.06- 6.94 (m, 12H, ArH), 3.30 (s, 6H, CH3). UV–Vis spectra [λmax, nm (log ε, L mol−1 cm−1); DMSO solution]: 265 (4.65), 355 (4.31), 400 (4.40), 425 (4.38), 465 (4.23), 600 (4.71). Anal. Calcd. for C28H20N4O3SZn: C, 60.2; H, 3.6; N, 10.0%. Found: C, 60.0; H, 3.3; N, 9.8%.
X-ray structure determination
Suitable crystals for single-crystal X-ray structure analysis were selected in mineral oil and mounted on glass fibers. Diffraction data for [Ni(L1)], [Ni(L2)] and [Zn(L3)(DMSO)] were collected on an IPDS-1 diffractometer (Stoe&Cie GmbH, Darmstadt, Germany) using graphite-monochromatized Mo-Kα radiation, (λ = 0.71073 Å). The structures were solved by direct methods using SHELXS and refined using SHELXL [17]. All non-hydrogen atoms with the exception of the disordered S atom of the DMSO ligand in [Zn(L3)(DMSO)] were refined anisotropically. Hydrogen atoms were included in idealized positions. The molecular graphics were drawn with DIAMOND [18]. Crystallographic data and details of the data collection and structure refinement are listed in Table 1, selected bond lengths and angles are presented in Table 2, and Scheme 3 shows packing views of the three complexes.
Microorganism strains and culture
In this research, we used one yeast (Candida albicans PTCC 5027), five standard bacteria (Pseudomonas aeruginosa ATCC 27853, Staphylococcus aureus PTCC 1112, Escherichia coli PTCC 1330, Bacillus cereus PTCC 1015 and Micrococcus luteus PTCC 1110) and three bacteria isolated from clinical samples (Klebsiella pneumoniae, Enterococcus faecalis and Listeria monocytogenes). Brain Heart Infusion medium (BHI-Merck) was used for L. monocytogenes and E. faecalis, whereas the other bacteria were cultured on Mueller–Hinton medium (Merck). Also, YGC medium (Merck) was used as the test medium for the yeast strain.
Determination of antimicrobial activity
The antimicrobial activities of [Ni(L1)], [Ni(L2)] and [Zn(L3)(DMSO)] complexes, NiCl2·6H2O and Zn(CH3CO2)2·2H2O against some Gram positive bacteria strains (L. monocytogenes, E. faecalis, S. aureus PTCC 1112, M. luteus PTCC 1110 and B. cereus PTCC 1015), some Gram negative bacterial strains (E. coli PTCC 1330, K. pneumoniae, P. aeruginosa ATCC 27853) and a fungal species (C. albicans PTCC 5027) were investigated using agar well diffusion plate methods [19]. The agar media were inoculated with 100 µl of the inoculums which were prepared using an overnight culture of each microorganism (18–24 h) adjusted to a turbidity equivalent to a 0.5-McFarland standard. Wells were cut and 50 µl of the compounds (10 mg/ml; DMSO was used as solvent) were added. Each compound was tested in triplicate along with standard ciprofloxacin for bacteria and fluconazole for yeast. The plates were incubated at 37 °C for 24 h. The antimicrobial activity was assayed by measuring the diameter of the inhibition zone formed around the well. The diameter of the zone of inhibition was measured by measuring scale in millimeters (mm). DMSO as solvent was used as a negative control. The minimum inhibitory concentration (MIC) and minimal bactericidal concentration (MBC) were determined by microdilution assay (NCCLS, 2008). The cultures were prepared in 24 and 72 h broth cultures of microorganisms, respectively. The MIC was defined as the lowest concentration of compound to inhibit the growth of microorganisms, and the MBC was defined as the lowest concentration of compound to kill the microorganisms. Serial dilutions ranging from 10 mg/ml to 39 µg/ml were prepared in medium.
Results and discussion
Syntheses and spectroscopic characterization
In general, metal complexes [Ni(L1)], [Ni(L2)] and [Zn(L3)(DMSO)] were obtained from the condensation of DAMN with various aldehydes in the presence of metal atoms. The IR spectra of the complexes exhibit absorption bands at 1612, 1611 and 1601 cm−1 respectively, which are assigned to the (C=N) stretching vibration [20]. The absence of an OH stretching vibration suggests that the O phenolic atoms of the ligands are deprotonated and participate in coordination to the nickel and zinc atoms [21]. The symmetric diimine nature of the complexes is proven by the single band observed in the range of 2209–2225 cm−1 assigned to the (C≡N) stretching vibration, contrary to two bands observed in monoimine compounds [22]. The absorption bands in the range of 735–754 cm−1 and 412–418 cm−1 in the metal complexes are assigned to stretching modes of the M–N and M–O bonds, respectively [23]. In the 1H NMR spectra of the complexes, the signal at 9.25, 8.57 and 8.49 ppm respectively is attributed to azomethine protons [15]. The electronic spectra of the complexes were recorded in DMSO. The bands at 265 and 270 nm are attributed to the electronic transitions π→π* of the aromatic rings. The bands between 305 and 400 nm are due to n→π* transitions. The bands above 400 nm are attributed to the intense charge transfer and intraligand transitions, indicating efficient conjugation in the metal complexes [24,25,26].
Structure description of [Ni(L1)] and [Ni(L2)]
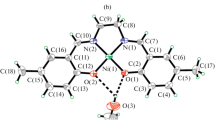
The molecular structure of [Ni(L1)] in solid state is shown in Fig. 1. This complex crystallizes in the orthorhombic space group Pbcn. The Schiff base ligand coordinates to one nickel atom in a tetradentate manner via phenolate O and imine N atoms. The geometry around the nickel atom is square planar, and the angle sum in the nickel plane is 360.1°. The bond distances between Ni(1) and O are 1.836(5) and 1.834(5) Å, and the Ni(1)–N bond lengths are 1.854(6) and 1.862(6) Å [27].
The molecular structure of [Ni(L2)] in solid state is shown in Fig. 2. [Ni(L2)] crystallizes in the monoclinic space group I2/a. The nickel atom is also four-coordinated with a square planar geometry. The coordination environment of the nickel atom is very similar to that of the [Ni(L1)].
Structure description of [Zn(L3)(DMSO)]
The molecular structure of [Zn(L3)(DMSO)] in the solid state is shown in Fig. 3. This complex crystallizes in the triclinic space group P\( \bar{1} \). The zinc atom is surrounded by two nitrogen atoms and two oxygen atoms from the Schiff base ligand and one oxygen atom from a DMSO ligand. The zinc atom is five-coordinated with a square pyramidal coordination geometry. The tetradentate ligand occupies the equatorial plane, and one O-coordinating DMSO ligand (sulfur atom disordered on two positions with 87.8(2) and 12.2(2)%) occupies the apical position. The coordinating N and O atoms form a plane with a maximum deviation of 0.1254(9) Å. Zn(1) is positioned 0.4381(9) Å below this plane (Fig. 3), the angle sum in the zinc plane amounts to 349.9°. The average bond length of Zn–N is 2.0902 Å. The Zn(1)–O(3) bond distance [2.0430(16) Å] to the DMSO ligand is slightly longer than those of Zn(1)–O(1) and Zn(1)–O(2) to the tetradentate ligand (av, 1.9636 Å) [28,29,30].
Antimicrobial activities
[Ni(L1)], [Ni(L2)] and [Zn(L3)(DMSO)] as well as NiCl2·6H2O and Zn(CH3CO2)2·2H2O were tested for their in vitro antimicrobial activities (Table 3). Results showed that all three complexes have potential as antimicrobial agents. The complex [Ni(L1)] has activity against three Gram positive bacteria. The rest of the compounds show no selectivity between Gram positive and Gram negative bacterial strains. NiCl2·6H2O and complex [Ni(L2)] have an antifungal effect against Candida albicans PTCC 5027. As shown in Table 3, while E. faecalis and L. monocytogenes isolated from clinical samples are resistant to the antibiotics ciprofloxacin and fluconazole, E. faecalis is sensitive to [Ni(L1)], [Ni(L2)], NiCl2·6H2O and Zn(CH3CO2)2·2H2O and L. monocytogenes is sensitive to [Ni(L2)], [Zn(L3)(DMSO)], NiCl2·6H2O and Zn(CH3CO2)2·2H2O. The results of minimum inhibitory concentration (MIC) and minimum lethal concentration (MBC) experiments are shown in Table 4. As can be seen, the MIC and MBC values for [Ni(L1)] and [Ni(L2)] are considerably lower than that for NiCl2·6H2O and also, for [Zn(L3)(DMSO)] is lower than that for Zn(CH3CO2)2·2H2O. It means that low concentrations of the complexes not only inhibit the growth of microorganisms, but also kill them. So, they have both bacteriostatic and bactericidal effects.
Conclusion
In conclusion, three metal Schiff base complexes have been synthesized and structurally characterized. The results indicated that the ligands were N2O2-tetradentate coordinated to the metal atoms through both the azomethine N atoms and phenolic O atoms. The nickel(II) complexes had four-coordinate square planar geometries, while the zinc(II) complex was five-coordinated with square pyramidal coordination geometry. In the zinc(II) complex, the tetradentate ligand occupied the equatorial plane and one O-coordinating DMSO ligand occupied the apical position. The antimicrobial activities of the three complexes and NiCl2·6H2O and Zn (CH3CO2)2·2H2O were also studied against different microorganisms. The results showed that all three complexes have some potential as antimicrobial agents. Also, according to the results of MIC and MBC, these complexes have both bacteriostatic and bactericidal effects. Further studies on the antimicrobial activities of diaminomaleonitrile-based complexes are planned for the future.
References
Ray MS, Chattopadhyay Sh, Drew MGB, Figuerola A, Ribas J, Diaz C, Ghosh A (2005) Eur J Inorg Chem 2005:4562–4571
Costes JP, Dahan F, Laurent JP (1986) Inorg Chem 25:413–416
Bian HD, Xu JY, Gu W, Yan SP, Cheng P, Liao DZ, Jiang ZH (2003) Polyhedron 22:2927–2932
Losada J, Del Peso I, Beyer L (2001) Inorg Chim Acta 321:107–115
Santos MLP, Bagatin IA, Pereira EM, Ferreira AMDC (2001) J Chem Soc Dalton Trans 6:838–844
Reddy PS, Ananthalakshmi PV, Jayatyagaraju V (2011) Eur J Chem 8:415–420
Ran XG, Wang LY, Lin YC, Hao J, Cao DR (2010) Appl Organomet Chem 24:741–747
Orio M, Jarjayes O, Kanso H, Philouze C, Neese F, Thomas F (2010) Angew Chem Int Ed 49:4989–4992
Fleck M, Layek M, Saha R, Bandyopadhyay D (2013) Transit Metal Chem 38:715–724
Raman N, Kulandaisamy A, Thangaraja C, Jeyasubramanian K (2003) Transit Metal Chem 28:29–36
Viswanathamurthi P, Natarajan K (1999) Transit Metal Chem 24:638–641
Bendre RS, Tadavi SK, Patil MM (2018) Transit Metal Chem 43:83–89
Al-Azmi A, Elassar AZA, Booth BL (2003) Tetrahedron 59:2749–2763
MacLachlan MJ, Park MK, Thompson LK (1996) Inorg Chem 35:5492–5499
Sheikhshoaie I, Ebrahimipour SY, Lotfi N, Mague JT, Khaleghi M (2016) Inorg Chim Acta 442:151–157
Lotfi N, Sheikhshoaei I, Ebrahimipour SY, Krautscheid H (2017) J Mol Struct 1149:432–438
Sheldrick GM (2008) Acta Crystallogr Sect A Found Crystallogr 64:112–122
Brandenburg K (2010) DIAMOND Version 3.2f Crystal Impact GbR Bonn, Germany
Irshad S, Mahmood M, Perveen F (2012) Res J Biol 2:1–8
Kumar V, Mishra RK, Shukla S, Mishra R, Singh M, Tiwari I, Thapliyal K, Upadhyay KK (2013) J Mol Struct 1047:66–72
Ebrahimipour SY, Ranjabr ZR, Kermani ET, Amiri BP, Rudbari HA, Saccá A, Hoseinzade F (2015) Transit Metal Chem 40:39–45
Lacroix PG, Di Bella S, Ledoux I (1996) Chem Mater 8:541–545
Tolulope MF, Olorunfemi O (2014) Der Pharma Chem 6:18–22
Gehad G, Abd El-Wahab MZ (2005) Spectrochim Acta 61:1059–1068
Wang P, Hong Z, Xie Z, Tong Sh, Wong O, Lee CS, Wong N, Hung L, Lee Sh (2003) Chem Commun 14:1664–1665
Ledoux I, Zyss J (1996) Pure Appl Opt 5:603–612
Costes JP, Lamère JF, Lepetit Ch, Lacroix PG, Dahan F (2005) Inorg Chem 44:1973–1982
Aazam ES, Ng SW, Tiekink ERT (2011) Acta Cryst E 67:m314–m315
Wang ZCh, Chu J, Zhan ShZ (2013) Acta Cryst E 69:m419
Lin ChW, Chou PT, Liao YH, Lin YCh, Chen ChT, Chen YCh, Lai ChH, Chen BS, Liu YH, Wang ChCh, Ho ML (2010) Chem Eur J 16:3770–3782
Acknowledgements
The authors gratefully acknowledge the financial support provided for this work by the Shahid Bahonar University of Kerman and also by the Universität of Leipzig, Germany, 04103 Leipzig, Germany.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Sheikhshoaie, I., Lotfi, N., Sieler, J. et al. Synthesis, structures and antimicrobial activities of nickel(II) and zinc(II) diaminomaleonitrile-based complexes. Transit Met Chem 43, 555–562 (2018). https://doi.org/10.1007/s11243-018-0241-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11243-018-0241-5