Abstract
A Schiff base derived from naringenin (NTSC) and its complex with Cu(II) ([Cu(H3L)(OAc)]·H2O, Cu(II)–NTSC) have been synthesized and characterized by physicochemical and spectroscopic methods. EPR studies confirmed that nitrogen, oxygen, and sulfur are the donor atoms bound to Cu(II) in the complex. The geometry of the complex has been modelled using DFT methods. Furthermore, naringenin and NTSC were used for the formation of Cu(II) complexes in solution, for comparison of biological activities. Antioxidant studies confirmed better radical scavenging activity of both NTSC and its Cu(II) complex compared to naringenin. The interaction of these compounds with calf thymus DNA was monitored by UV–Vis spectroscopy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Natural flavonoids present in fruits and plant-derived foods are known to have a number of health-promoting effects, as confirmed by many experimental and epidemiological studies [1]. In particular, the promising bioactivities of flavanones, a subclass of flavonoids, and their impact on human health are of interest. Flavanones can exhibit antioxidant, immunomodulatory, chemopreventive, and anticancer properties [2].
On the other hand, thiosemicarbazides are versatile ligands characterized by the π delocalization of charge and the configuration flexibility of the molecular chain, which can give rise to a great variety of coordination modes. These abilities have a great impact on the biological properties of thiosemicarbazides, which include: anticancer, antifungal, antibacterial, antiviral, and antimalarial properties [3]. These observations have prompted us to synthesize a novel naringenin thiosemicarbazone Schiff base in order to obtain a modified flavanone combining the physicochemical properties of thiosemicarbazides and the biological properties of flavanones.
A wide range of Schiff bases with reactive azomethine linkages show interesting inhibitory activity against tumor cells [4]. Moreover, the use of metal complexes as therapeutic drugs is very promising. Copper is a well-known bioessential element, and its complexes have been found to be appropriate for biological applications due to their binding ability and positive redox potential. Cu(II) complexes of heterocyclic bases have extensively been explored by virtue of their strong interactions with DNA and cytotoxic activities. In addition, they can act as artificial nucleases. It is commonly known that Cu(II) complexes demonstrate an efficient DNA cleavage activity [5, 6]. Thus, their cytotoxicity may be caused by their ability to bind and cleave DNA, leading to cell cycle arrest and apoptosis or generation of reactive oxygen species (ROS) which in turn it leads to cell death. Furthermore, some Cu(II) complexes of Schiff bases show antibacterial and antiproliferative activity due to the properties of the metal center or the coordinated ligands alone, as well as the structural and electronic properties which result from coordination [7].
To date, Schiff base ligands based on flavonoids and thiosemicarbazides have not extensively been investigated. The aim of this study was to obtain a novel naringenin derivative and determine the impact of structural modification of the molecule on its antioxidant activity, acid–base profile, chelating ability, and interactions with calf thymus (CT) DNA. The copper(II) complex of this ligand has been prepared and characterized.
Experimental
Materials and instrumentation
5,7-Dihydroxy-2-(4-hydroxyphenyl)chroman-4-one (racemic naringenin) (98 %), DMSO, 2,2-diphenyl-1-picrylhydrazyl (DPPH), 2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS) (≥99.0 %), (±)-6-hydroxy-2,5,7,8-tetramethylchroman-2-carboxylic acid (Trolox), 2,4,6-tri(2-pyridyl)-s-triazine (TPTZ), and iron(II) sulfate heptahydrate were purchased from Sigma-Aldrich. Methanol (99.8 %) was purchased from Avantor Performance Materials Poland S.A. (Gliwice, Poland). Sodium hydroxide and potassium hydrogen phthalate (KHP) were received from Merck. Potassium nitrate was purchased from Sigma-Aldrich. A carbonate-free sodium hydroxide solution was prepared and standardized against pure potassium hydrogen phthalate (KHP) and then used in spectrophotometric titrations.
Elemental analysis (C, H, and N) were carried out using a EuroVector 3018 analyzer. The metal content of the complex was determined using AAS GBC 932 Plus (Australia) with a copper hollow-cathode lamp. Melting points were measured with an Electrothermal 9200 microscopic melting point apparatus. IR spectra were recorded employing a Nicolet 6700 FTIR spectrometer (Thermo Elektron Corp.), in the 4000–400 cm−1 region. KBr disks for FTIR analysis were performed using a Specac hydraulic press. 1H NMR spectra were recorded on a Bruker AV200 200-MHz spectrometer in DMSO-d6 with TMS (tetramethylsilane) as internal standard. EPR spectra were recorded in the solid state and in DMF solution from 0 to 8000 Gauss at 100 K with an X-band (9.4-GHz) Bruker EMX spectrometer equipped with an HP 53150A microwave frequency counter. Mass spectra were obtained on a Finnigan MAT 9 instrument. Antioxidant and CT-DNA measurements were taken on a Hewlett-Packard 8453 spectrophotometer. Spectrophotometric studies were carried out using a UV light beam generator connected to a potentiometric apparatus to dose the appropriate volume of sodium base. Titrations were conducted using an automatically set Molspin pHmeter (Molspin Ltd., Newcastle-upon-Tyne, UK) equipped with a micro-Molspin DSI 0.250 mL controlled by a PC. The measuring electrode was a microelectrode Russell CMAWL/S7. Absorbance was measured using Ocean Optics software.
Synthesis of NTSC
The synthesis of the ligand is shown in Scheme 1. Naringenin (312.65 mg, 1.2 mmol) and thiosemicarbazide (130.49 mg, 1.4 mmol) were dissolved in ethanol (10 ml). Concentrated H2SO4 (50 µl) was added to this solution. The reaction mixture was refluxed at 60 °C for about 24 h with stirring. The resulting dark yellow solution was concentrated and cooled, giving a light yellow crystalline precipitate. This was filtered off, washed with distilled water and ethanol, and recrystallized from DMF and water to give NTSC. Yield: 328.48 mg (87 %). 1H NMR (DMSO-d6, 200 MHz), δ (ppm) 2.75–3.09 (m, 2H, CH2); 5.02 (d, 1H, 3JH–H = 9.1 Hz, CH); 5.81–5.87 (m, 2H, 2 × CH aromatic); 6.76 (d, 2H, 3JH–H = 8.4 Hz, 2 × CH aromatic); 7.27 (d, 2H 3JH–H = 8.4 Hz, 2 × CH aromatic); 9.53 (s, 2H, NH2); 9.91 (bs, 1H, NH); 10.53 (bs, 3H, OH). M.p. 268–270.5 °C. Anal. Calc. for C16H15N3O4S: C 55.6 %; H 4.4 %; N 12.2 %; S 9.3 %. Found: C 55.3 %; H 4.4 %; N 11.2 %; S 9.6 %. IR ν max(cm−1): ν(N–H): 3438–3264–3168, ν(Ar–OH): 3354, ν(O–H): 3293, 3427, ν(C=N): 1636, ν(C=C): 1517–1503–1477, ν(C–O): 1344, ν(C–O–C): 1152, ν(C–N): 1066, ν(C=S): 826. UV–Vis λ max(nm): 277, 332 nm.
Synthesis of the complex
The complex was prepared in accordance with the procedure described by Li and Yang [8] but with some modifications. NTSC (51.32 mg, 0.1 mmol) was dissolved in acetone (10 ml). After 5 min, Cu(OAc)2·H2O (38.42 mg, 0.2 mmol) was quickly added to the solution, which was then refluxed at 70 °C for about 5 h with stirring. The resulting dark brown precipitate of [Cu(H3L)(OAc)]·H2O was separated from the solution by filtration, washed with ethanol, and dried in vacuo. 1H NMR analysis of the complex was not possible due to the lack of resolution of the spectra for the paramagnetic nature of Cu(II) atom [9, 10]. Yield: 37.54 mg (37 %). M.p. 210 °C. Anal. Calc. for [Cu(H3L)(OAc)]·H2O: C 42.6 %; H 3.9 %; N 8.7 %; S 6.6 %; Cu 13.0 %. Found: C 42.3 %; H 3.5 %; N 9.0 %; S 6.8 %; Cu 12.8 %. IR ν max(cm−1): ν(O–H): 3325, ν(Ar–OH): 3317, ν(N–H): 3192, ν as(COO−): 1548, ν s(COO−): 1358, ν(C=N): 1593, ν(C=C): 1517–1503–1477, ν(C=S): 827, ν(M–N):411, ν(M–S):430 and ν(M–O):443. UV–Vis λ max(nm): 270, 340, 363 nm.
DFT calculations
DFT (density functional theory) calculations were performed with Gaussian 03 (revision C.02) software. The geometry of the Cu(II) complex was optimized with Gaussian using the B3LYP functional and 6-311+g(d) basis set on S plus 6-311g on all the other atoms, by the procedure reported in the literature [11]. This choice generally ensures a good degree of accuracy in the prediction of the structures of first-row transition metal complexes. For all the structures, minima were verified through frequency calculations.
The ORCA program was used to calculate the g and A tensors of the 63Cu center in this complex. The method for calculation of g and A tensors was developed and implemented into the ORCA package by Neese [12]. The functionals used were B3LYP and B3PW91 and the basis set VTZ, according to the procedure established in the literature [13]. The 63Cu hyperfine coupling tensor A has three contributions: the isotropic Fermi contact (AFC), the anisotropic or dipolar hyperfine interaction (AD), and one second-order term that arises from spin-orbit (SO) coupling (ASO): \( \varvec{A} = \, A^{{\text{FC}}} {\mathbf{1 }} + \varvec{A}^{\text{D}} + \, \varvec{A}^{{\text{SO}}} \), where 1 is the unit tensor. The values of the 63Cu anisotropic hyperfine coupling constants along the x, y, and z axes are as follows: \( A_{\text{x}} = \, A^{{\text{FC}}} +A_{\text{x}}^{\text{D}} + \, A_{\text{x}}^{{\text{SO}}} \), \( A_{\text{y}} = \, A^{{\text{FC}}} +A_{\text{y}}^{\text{D}} + \, A_{\text{y}}^{{\text{SO}}} \), and \( A_{\text{z}} = \, A^{{\text{FC}}} +A_{\text{z}}^{\text{D}} + \, A_{\text{z}}^{{\text{SO}}} \). The theory background has been described in the literature [14]. The percent deviation from the experimental values of gi and |A i | (i = x, y, z) was calculated as 100 × [(|g i |calcd − |g i |exptl)/|g i |exptl] and 100 × [(|A i |calcd − |A i |exptl)/|A i |exptl].
DPPH radical scavenging activity
DPPH assays were done in accordance with the modified method of Brand-Williams et al. [15]. The results are expressed as µM Trolox equivalent (TE). Determinations were carried out in triplicate, and the results are shown as mean values ± SD.
ABTS+ radical scavenging activity
Total antioxidant capacity assays were carried out using the modified ABTS+ method described by Re et al. [16] and by Cai et al. [17]. Three independent experiments were carried out for each of the samples, and the results are expressed as Trolox equivalent antioxidant capacity (TEAC, µM Trolox equivalent of studied sample).
Ferric-reducing antioxidant power assay (FRAP)
Ferric-reducing ability was measured according to the procedure of Benzie and Strain [18] with some modifications (concentrations and volumes of the samples). Experiments were performed in triplicate, and the results are expressed as mM FeSO4·7H2O equivalent.
Acid–base characteristic determination
The acid–base profiles were determined using apparatus for potentiometric titration connected to a UV light beam generator by a fiber optic cable. The investigations were conducted for naringenin, compounds previously synthesized (NTSC, Cu–NTSC), and compounds not isolated but formed in solution only (Nar/Cu(II) and NTSC/Cu(II)) using different ligand-to-metal (L/M) ratios: (1/0), (1/1), (1/2). All samples were prepared in DMSO solvent and diluted with HNO3 (6.25 mM, H2O) possessing constant ionic strength of 0.1 M by virtue of KNO3 present in the solution. Titrations of tested compound solutions (30 µM, 1.2 % DMSO/98.8 % H2O) were carried out in the pH range from 2.5 to 12.5 using 0.1 M NaOH as a titrant.
Interactions with calf thymus DNA
Spectrophotometric titrations were carried out by stepwise additions of calf thymus (CT) DNA solution (0.46 mM, in 5 mM Tris–HCl, pH 7.2, and 50 mM NaCl buffer) to a solution of each compound (0.25 µM) in DMSO. The UV–Vis spectrum was recorded after each addition. The intrinsic binding constants between the analyzed molecules and DNA were calculated [19]. The intrinsic DNA binding constants K b for Nar, NTSC, and their complexes with Cu(II) ions, and the Cu–NTSC complex were determined according to the following equation:
where [DNA] is the concentration of DNA in base pairs; ɛ a, ɛ f, and ɛ b are the apparent, free, and bound complex extinction coefficients, respectively. The value of ɛ f was calculated according to Beer’s law; ɛ a was determined as the ratio between the measured absorbance and the concentration of each tested compound, A obs/c compound. A plot of [DNA]/(ɛ a − ɛ f) versus [DNA] gave a straight line with a slope of 1/(ɛ b − ɛ f) and a y-intercept equal to 1/K b (ɛ b − ɛ f); K b is the ratio of the slope to the y-intercept.
Spectrophotometric titrations of CT-DNA with the tested compounds (Nar, NTSC, their complexes with copper “in situ”, and the Cu–NTSC complex in solid state) were done in order to gain further insights into the mechanism of interaction of these compounds with CT-DNA [20]. The titrations were carried out by stepwise addition of each compound solution (2.5 mM) to a solution of CT-DNA (0.25 µM).
Statistical analysis
All determinations were carried out in triplicate. Mean values with standard deviations (±SD) are reported for each case. Significant differences for multiple comparisons were considered by t test for dependent samples (Statistica 10) at the p < 0.05 level. All obtained spectra were analyzed using OriginPro 8.0 (OriginLab, USA) software.
Results and discussion
Characterization of NTSC and its copper(II) complex
Both NTSC and its copper(II) complex were air-stable for prolonged periods and exhibited good solubility in DMSO and DMF, but were insoluble in acetone, chloroform, ethanol, methanol, and water. The UV–Vis electronic spectra of free NTSC and the complex were recorded in DMSO solution. Free NTSC has an intense band at 332 nm and a less intense band at 277 nm. The complex [Cu(H3L)(OAc)]·H2O gives bands at 270, 340, and 363 nm. The bands at 450 (sh) and 635 nm correspond to a CT (charge transfer transition) S → Cu and d–d transition of Cu(II) coordinated by an {O, N, S} donor atom set, respectively (see Supplementary material Fig. S1). In the FTIR spectrum of free NTSC, peaks at 3293 and 3427 cm−1 are assigned to the hydroxyl group. Peaks at 1636, 1517–1477, and 826 cm−1 are assigned to C=N, C=C, and C=S, respectively. In the FTIR spectrum of [Cu(H3L)(OAc)]·H2O, peaks at 3325, 1593, 1517–1477, and 827 cm−1 are assigned to OH, C=N, C=C, and C=S, respectively. A strong band at 1636 cm−1 due to C=N in the free ligand is shifted to 1593 cm−1 in the spectrum of the complex (Fig. S2). This shift to a lower frequency region is associated with the decrease in electron density in the azomethine linkage, consistent with coordination of this group to the metal center [21], as well as through the oxygen atom of the acetate group. Bands at 411, 430, and 443 cm−1 are attributable to ν(M–N), ν(M–S), and ν(M–O), respectively. The lower intensity and the shift in frequency corresponding to ν(C=S) confirmed that the complex is formed via conversion of the C=S group to thiolate, indicating that NTSC can act as a monobasic tridentate ligand [22]. The Cu–O and Cu–N stretching vibrations are expected in the range of 600–100 cm−1, as reported in the literature [23]. [Cu(H3L)(OAc)]·H2O displays asymmetric as well as symmetric stretching COO− vibrations at 1548 and 1358 cm−1, respectively. The Δν = 190 cm−1 is bigger than that of sodium acetate (153 cm−1), indicating that the acetate group is monodentate in this complex.
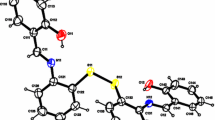
The proposed geometry of Cu(II)–NTSC has been optimized by DFT according to the procedure established in the literature [11], and the resulting optimized structure is shown in Fig. 1. It is a slightly distorted square planar geometry, which is usual for copper(II) complexes. The calculated bond lengths and angles fall in the expected ranges (Table 1) [24].
Structure of the Cu(II)–NTSC complex optimized by DFT calculations. Definition of atom color: red oxygen, blue nitrogen, orange copper, gray carbon, white hydrogen (see “Experimental” section for details of the procedure). (Color figure online)
The X-band EPR spectrum recorded on the polycrystalline solid complex at 100 K is characterized by two g values (g || = 2.191 and g ⊥ = 2.053) with the order g || > g ⊥ > g e (Fig. 2). These observations suggest an S = 1/2 system with axial symmetry and a \( d_{{x^{2} - y^{2} }} \) ground state, indicative of a square planar geometry [25]. When the solid compound is dissolved in DMF, four resonances in the parallel region of the spectrum are observed, due to coupling between the unpaired electron and 63,65Cu nucleus with I = 3/2 (Fig. 3). The spectrum is characterized by g || = 2.180, A || = 185.0 × 10−4 cm−1 and g ⊥ = 2.040, A ⊥ = 25.0 × 10−4 cm−1. The values of g || and A || appear to be compatible with a Cu(NO2S) coordination environment. In the first three parallel resonances, the superhyperfine coupling between the unpaired electron and the 14N nucleus (I = 1) is observable; for each resonance, this coupling results in two different triplets with intensity 1:1:1, due to the presence of 63Cu (69.1 %) and 65Cu (30.9 %) isotopes in the natural copper, which overlap partially (this is evident mainly in the lowest field resonance). This confirms that only one nitrogen donor is bound to copper, in agreement with the structure shown in Fig. 1. An analogous coupling with 14N was recently observed for a Cu(II) complex of a hesperetin hydrazone ligand [5]. The value of 16.8 × 10−4 cm−1 for A N|| is in good agreement with those reported in the literature for other Cu(II) species [26].
To confirm the EPR results, DFT calculations were carried out on the complex using ORCA software, in order to obtain the hyperfine coupling tensor A (these calculations need to include appropriate spin polarization which can be achieved through the use of the UKS formalism; moreover, the spin-orbit contribution to A, calculable by ORCA, was found to become more and more important for late 3d elements, in particular Cu [12], and its inclusion in the calculations seems essential to reach a good agreement with the experimental data). The calculated g || and A || values are compared with the experimental ones in Table 2. Recently, the prediction of EPR parameters (in particular A ||) was validated for the species formed by simple oligopeptides and it was found that the B3LYP and B3PW91 functionals give a good agreement with the experimental data [13]. In particular, B3PW91 slightly overestimates A || and B3LYP slightly underestimates A ||; these calculations also allow one to predict the value of g ||. Based on this observation, EPR parameters for the present complex were calculated with B3LYP and B3PW91 functionals and the VTZ basis set. The results are reported in Table 2. It is noteworthy that the experimental value of A || (which in this case coincides with A z ) falls between those calculated with B3PW91 and B3LYP, suggesting that the proposed structure for the complex is correct. As shown in Figs. 2 and 3, the calculated anisotropy between the x and y axes is negligible (thus A x ~ A y = A ⊥).
Antioxidant activity
Antioxidant activity studies were carried out for the synthesized compounds (NTSC and Cu(II)–NTSC) as well as for naringenin and also the non-isolated complexes (Nar/Cu(II), NTSC/Cu(II)) using different ligand-to-metal ratios. Two different solvents (DMSO and methanol) were used for naringenin only, for comparison. The inhibitory effects of the compounds on the DPPH radical are given in Table 3. The average scavenging ratio for DPPH increases with amounts of the compound. The TEAC (Trolox equivalent antioxidant capacity) value calculated for DPPH presents the highest scavenging power for the highest concentration tested (100 µM). Significant scavenging capacity is observed for NTSC (99.13 ± 1.31 µmol/L). Furthermore, the complex of NTSC with copper(II) (1/1 v/v) (102.19 ± 1.58 µmol/L) possesses greater antioxidant activity than complexes of naringenin with Cu(II) (1/1 v/v) in DMSO and methanol solvents. The IC50 value, defined as the concentration of the sample required for 50 % scavenging of DPPH free radicals, was also calculated. A smaller IC50 value corresponds to greater potency. The IC50 values (Table 3) indicate that NTSC (0.43 mM) and its complex with Cu(II) in ratio 1/1 (0.43 mM) are the most effective DPPH radical inhibitors, followed by the other naringenin derivative complexes. The average inhibitory ratio of the free ligand (IC50 = 9.99 mM; IC50 = 9.23 mM) is the least for naringenin. Nguyen et al. [27] showed that IC50 values for a series of thiosemicarbazone compounds obtained from the condensation reaction of N-(tetra-O-acetyl-β-d-galactopyranosyl)thiosemicarbazide with a number of substituted benzaldehydes were slightly lower than IC50 values in this study. The scavenging activity of the thiosemicarbazones (NTSC, Cu(II)–NTSC, NTSC/Cu = 1/1, NTSC/Cu = 1/2) can be attributed to the N–H group in the thiosemicarbazone moiety, which can donate a hydrogen atom to the DPPH radical. The resulting free radicals are stabilized by delocalization into the benzene ring. When the concentration of thiosemicarbazone is higher than that of DPPH radical, the DPPH radical is completely consumed; at lower concentrations, the residual DPPH radical may react with the resulting thiosemicarbazone radicals [15, 27]. The choice of solvent shows no significant influence on the DPPH scavenging ability.
The total antioxidant capacities (TEACABTS) of the tested compounds (Table 3) show a wide variation. TEAC values are the highest for the highest concentration (100 μM) of all compounds. Free naringenin in DMSO solution (71.25 ± 12.03 µmol/L) is a better ABTS radical cation inhibitor than in methanol (24.29 ± 3.36 µmol/L). Addition of excess copper(II) to naringenin solution leads to higher TEAC values in DMSO (40.18 ± 2.56 µmol/L) but lower in methanol (15.96 ± 2.26 µmol/L). The highest antioxidant capacity is assigned to Cu(II)–NTSC (99.43 ± 0.45 µmol/L). The IC50 value calculated for TEACABTS (Table 3) shows that the best ABTS radical cation inhibitors are the synthesized complex Cu(II)–NTSC (0.17 mM), as well as NTSC/Cu = 1/1 (0.19 mM), NTSC/Cu = 1/2 (0.20 mM), and NTSC (0.21 mM). The weakest antioxidant capacity is found for the naringenin complex in methanol (1/2 v/v). According to Re et al., naringenin has very similar Trolox equivalent values, which depend on the time [16]. In case of the thiosemicarbazones, it is observed that the antioxidant activity is the same for the free ligand and its metal complexes. In contrast, Yousef et al. [28] reported lower antioxidant capacities for metal complexes of thiosemicarbazones in comparison with the free ligand. The effective thiosemicarbazone inhibition can be attributed to the presence of the thiazole and phenolic moieties which are likely to be structural components contributing to ABTS radical cation scavenging activity. Furthermore, the presence of hydroxyl groups in the tested compounds increases the antioxidant and scavenging activities. The solvent shows a significant influence on ABTS radical cation scavenging activity; DMSO gives much higher antioxidant capacities than methanol. This can be explained by the scavenging properties of DMSO and its direct reaction with free radicals. The mechanism of ABTS+ action for the compounds in methanol may involve the dismutation of free radicals into compounds of lower activity.
In the present study, the results of ferric-reducing antioxidant power (FRAP) demonstrate no significant differences in the whole range of concentrations (Table 3). Nevertheless, a few significant FRAP values can be distinguished. The highest reducing power is shown for the Cu(II)–NTSC complex (7.35 ± 0.09 mM FeSO4), followed by NTSC/Cu = 1/1 (7.17 ± 0.09 mM FeSO4), NTSC (7.06 ± 0.05 mM FeSO4), and NTSC/Cu = 1/2 (6.84 ± 0.05 mM FeSO4). The IC50 for this assay shows that the complex Cu(II)–NTSC has the highest ability to reduce Fe3+ to Fe2+ (8.88 mM). NTSC and its complexes (1/1, 1/2 v/v) also possess good ferric-reducing power. In accordance with Firuzi et al. [29], naringenin displays lower FRAP values, which are also time dependent.
Acid–base profile determination
Spectrophotometric titrations of all these compounds were carried out in 1.2 % DMSO/98.8 % H2O solution. The UV–Vis spectrum of naringenin was recorded in DMSO. The positions and intensities of the bands provide valuable information concerning the type of flavonoid and their oxidizing nature. Figure S3 (Supplementary material) shows two major absorption bands in the UV–Vis region for naringenin. The band at 315–370 nm corresponds to the B and C ring portion (cinnamoyl system, band I), and the band at 260–310 nm is associated with the A and C ring portion (benzoyl system, band II). The shoulder at 333 nm corresponds to \( n {-} \pi ^{*} \) interactions, and the shoulder at 289 nm corresponds to \( \pi {-} \pi ^{*} \) interactions, and HOMO–LUMO transitions were also observed. The band at 258 nm is assigned to intra-ligand transitions [30].
The spectrum of naringenin showed a bathochromic shift with maximum at 323 nm with increasing pH, associated with the degree of ionization of the hydroxyl groups (Fig. S4A). This band is bathochromically shifted with a maximum at 420 nm in very strong alkaline conditions (Fig. S4B), under which chalconaringenin can be formed. Naringenin as a dihydroflavanone does not possess a double bond at C2 and C3 in its structure; hence, there is no conjugation of electron density between the phenol and benzoyl moieties [31]. The acid dissociation constants of the naringenin hydroxyl groups have been published in our previous study [32]. Band shifts in the pH range 2.24–14.0 allow estimation of the pK a constants for naringenin which are comparable with the literature data [31]. Due to the presence of a carbonyl group in the C ring and a negative mesomeric impact on the redistribution of the electron density in the A ring, an increase in the pK a for the OH group at the 7 position (para position) of the A ring is supposed to occur. It is likely that this group refers to the lowest value of the dissociation constant (pK a1), while the value of pK a2 corresponds to the hydroxyl group at the 4’ position of the B ring. The highest dissociation constant (pK a3) is assigned to the OH group at the 5 position, which is presumably involved in a hydrogen bond between the hydroxyl proton at the 5 position and the carbonyl at the 4 position [32].
The UV–Vis spectra of the naringenin complex with Cu(II) in ratio 1/1 (Fig. 4) show a bathochromic shift of band II with a maximum at 231 nm and a decrease in its intensity. A bathochromic shift with a maximum at 318 nm and an increase in intensity are observed for band I. However, the spectra of the complex of naringenin with excess copper(II) show only one bathochromic shift at 319 nm for band I and a decreased intensity of band II (Fig. S5). The lower intensity of band II for naringenin complexes can be explained by the formation of a new six-membered ring system between the metal and the oxygen atoms from the 4-oxo and 5-OH sites. The similarity for the spectra at different naringenin/copper ratios (1/1 and 1/2) confirms the same donor–acceptor systems for these two complexes.
The electronic absorption spectra of NTSC are characterized by various forms depending on the pH. Significant bathochromic shifts were observed for the naringenin derivative, as shown in Fig. S6. In alkaline medium, a new band is observed at around 385 nm and the intensities of the other bands decrease. The band at 385 nm is assigned to the \( n {-} \pi ^{*} \) transition of the azomethine group of the naringenin thiosemicarbazone molecule.
The UV–Vis spectrum of NTSC exhibits notable changes in the presence of copper(II), suggesting complexation. Similar observations have been reported in the literature [33]. In the spectrum of the complexes, the bands of the azomethine \( n {-} \pi ^{*} \) transition are shifted to lower frequencies, indicating that the imine nitrogen coordinates to the metal. The new band at 365 nm for all the NTSC/copper(II) mixtures under alkaline conditions is assigned to the thiosemicarbazone moiety (Fig. S7-9).
Interactions with CT-DNA
Electronic absorption spectroscopy is commonly used to study the interactions of compounds with DNA. Intercalative binding to DNA usually results in hypochromism and bathochromism because of π–π stacking interactions between the aromatic chromophore and the base pairs of DNA. The extent of hypochromism is generally correlated with the intercalative binding strength.
The absorption spectra of naringenin, NTSC, and its complexes with Cu(II) in the absence (spectrum at the bottom in Fig. S10 A–C and at the top in Fig. 5 and Fig. S10 D–H) and presence of CT-DNA are shown in Fig. 5 and Fig. S10. Titration of these compounds with increasing concentrations of CT-DNA results in changes in the spectra which show double hyperchromism in different bands, reaching 82.4 % at 253 nm and 3.5 % at 323 nm, 83.5 % at 251 nm and 2.9 % at 323 nm, 70.6 % at 251 nm and 2.3 % at 323 nm, for naringenin in DMSO solvent, naringenin in methanol, and the 1:1 complex of naringenin with Cu(II) in DMSO, respectively. The ratio of [DNA]/[compound] was 0.01. In addition, the hyperchromism observed for all three of those systems is accompanied by a small red shift of 2 to 4 nm and an increase in molar absorptivity, i.e., hyperchromism. Such changes are specific for compounds bound to double-stranded DNA through non-covalent interactions, such as electrostatic and van der Waals forces, dative bonds, hydrogen bonds, or hydrophobic interactions [34].
Electronic absorption (UV–Vis) spectra of Cu(II)–NTSC complex (a) and Nar/Cu = 1/2 in MeOH (b) in Tris–HCl buffer (pH 7.4) at room temperature in the presence of increasing amounts of CT-DNA. Arrows indicate the change in absorbance upon increasing DNA concentration. Inset plot of [DNA]/(ɛ a − ɛ f) versus [DNA]
On the other hand, other naringenin systems showed both hyperchromism and hypochromism with a slight red shift. Likewise, with increasing DNA concentrations, the absorption band at 251 nm for Nar/Cu = 1/1 in MeOH, Nar/Cu = 1/2 in DMSO, and Nar/Cu = 1/2 in MeOH shows hyperchromism of 91.7, 89.5, and 86.3 %, respectively. A small red shift of around 6 nm is also observed. According to Shahabadi and Mohammadi, this hyperchromic effect may be the result of electrostatic interactions between the positively charged [CuL2]2+ complex and the negatively charged phosphate backbone of the CT-DNA [35]. However, the band at 323 nm for Nar/Cu = 1/1 in methanol, Nar/Cu = 1/2 in DMSO, and Nar/Cu = 1/2 in methanol shows hypochromism of 21.8, 13.4, and 18.3 %, respectively.
NTSC and its complexes with Cu(II) showed a large hyperchromism with a slight red shift and a much smaller hypochromism. The band at 250 nm showed hyperchromism with 74.8 % for NTSC. The band at 257 nm showed hyperchromism with 56.1, 77.5, and 61.7 %, corresponding to NTSC/Cu = 1/1, NTSC/Cu = 1/2, and Cu(II)–NTSC. Moreover, the hypochromism is 5.6, 3.4, 4.9, and 19.9 % in relation to NTSC, Cu(II)–NTSC, NTSC/Cu = 1/1, and NTSC/Cu = 1/2, respectively. The absorption intensity increases (hyperchromism) with increasing CT-DNA concentration, suggesting degradation of the DNA double helix structure. Hypochromism results from the constriction of DNA helix axes and the conformational changes on the DNA molecule, while hyperchromism results from secondary damage to the DNA double helix structure. Additionally, a well-defined isosbestic point was observed at 301 nm for the complexes of naringenin and NTSC with excess copper in DMSO, followed by 306 nm for the complex of NTSC with copper (1/1). The presence of an isosbestic point indicates that there is only one mode of binding. For the other compounds, there is no clear isosbestic point, suggesting more than one way of binding with CT-DNA.
The intrinsic binding constants K b were calculated using Eq. (1). The highest K b value was found for the complex in solid-state Cu(II)–NTSC (3.0 × 106 M−1), followed by the complex of Nar/Cu(II) 1/2 in MeOH (2.0 × 106 M−1), NTSC (1.5 × 106 M−1), and the complex of naringenin with Cu(II) in ratio 1/1 in methanol (1.0 × 106 M−1). The weakest binding affinity was found for naringenin in DMSO (K b = 5.0 × 104 M−1) (Table 4). These values are slightly larger than those of well-known DNA intercalators, such as K b = 4.8 × 104 M−1 for [Ru(bpy)2(phi)]Cl2 (phi = 9,10-phenanthrenequinonediimine), 2.1 × 104 M−1 for [Ru(bpy)2(ddt)]2+ (ddt = 3-(pyrazin-yl)-5,6-diphenyl-as-triazine), and 6.3 × 104 M−1 for [Ru(bpy)2(dpt)]2+ (dpt = 3-(pyrazin-yl)-as-triazino[5,6-f]phenanthrene) [8]. The K b values show that the complexes bind to DNA more strongly than the free ligand.
In accordance with a competitive interaction of the test compounds and CT-DNA using solutions of the compounds as titrants, a large hyperchromism with a slight blue shift was observed in a few cases (Fig. S11 A–J). There is no visible isosbestic point in any of the spectra. Hyperchromism may indicate an electrostatic interaction with the double helix of DNA, perhaps involving the positively charged complex unit and the negatively charged phosphate DNA backbone [20, 35].
Conclusions
The thiosemicarbazone Schiff base derived from naringenin and its copper complex have been synthesized and characterized by several methods. The compounds demonstrate promising scavenging effects in DPPH, ABTS, and FRAP assays. The interaction of the ligand and complex with CT-DNA was investigated by absorption measurements. K b constant shows that the complex binds to CT-DNA more strongly than the free ligand. It confirmed that DNA could be an important target in cellular systems for this metal-based compound derived from flavonoid. The high binding affinity to DNA may have potential in drug development, but the detailed biological activities of these systems will require further investigation. Results obtained from present work will be useful to understand the mechanism of interaction of complexes with CT-DNA and helpful in the development of potential biological and physiological applications in the future.
References
Brodowska K, Łodyga-Chruścińska E (2014) Flavonoids and antioxidants. In: Govil JN, Bhattacharya S, Kaushik G (eds) Recent progress in medicinal plants, vol 40. Studium Press (India) Pvt. Ltd., New Delhi
Graf BA, Milbury PE, Blumberg JB (2005) J Med Food 8:281–290
Kalhor M, Shabani M, Nikokar I, Banisaeed SR (2015) Iran J Pharm Res 14(1):67–75
Kamel MM, Ali HI, Anwar MM, Mohameda NA, Soliman M (2010) Eur J Med Chem 45:572–580
Lodyga-Chruscinska E, Symonowicz M, Sykula A, Bujacz A, Garribba E, Rowinska-Zyrek M, Oldziej S, Klewicka E, Janicka M, Krolewska K, Cieslak M, Brodowska K, Chruscinski L (2015) J Inorg Biochem 143:34–47
Borowska J, Sierant M, Sochacka E, Sanna D, Lodyga-Chruscinska E (2015) J Biol Inorg Chem 20:989–1004
Abdi K, Hadadzadeh H, Weil M, Salimi M (2012) Polyhedron 31:638–648
Li Y, Yang Z-Y (2009) Inorg Chim Acta 362:4823–4831
Kozlevcar B, Segedin P (2008) Croat Chem Acta 81:369–379
Hathaway BJ (1987) In: Wilkinson G. In: Gillard RD, McCleverty JA (eds) Comprehensive coordination chemistry, vol 5. Pergamon Press, New York, pp 533–774
Micera G, Garribba E (2012) Int J Quantum Chem 112:2486–2498
Neese F (2013) ORCA—an ab initio, DFT and Semiempirical Program Package, Version 3.0, Max-Planck-Institute for Chemical Energy Conversion, Mülheim a. d. Ruhr
Kallay C, David A, Timari S, Nagy EM, Sanna D, Garribba E, Micera G, De Bona P, Pappalardo G, Rizzarelli E, Sovago I (2011) Dalton Trans 40:9711–9721
Micera G, Garribba E (2011) J Comput Chem 32:2822–2835
Brand-Williams W, Cuvelier ME, Berset C (1995) Lebensm Wiss Technol 28:25–30
Re R, Pellegrini N, Proteggente A, Pannala A, Yang M, Rice-Evans C (1999) Free Radical Biol Med 26:1231–1237
Cai Y, Luo Q, Sun M, Corke H (2004) Life Sci 74:2157–2184
Benzie IF, Strain JJ (1996) Anal Biochem 239:70–76
Wu JZ, Yuan L, Wu JF (2005) J Inorg Biochem 99:2211–2216
Ensafi AA, Hajian R, Ebrahimi S (2009) J Braz Chem Soc 20:266–276
Mendes IC, Teixeira LR, Lima R, Carneiro TG, Beraldo H (1999) Transit Met Chem 24:617–621
Rapheal PF, Manoj E, Prathapachandra Kurup MR, Suresh E (2007) Polyhedron 26:607–616
Helios K, Wysokiński R, Pietraszko A, Michalska D (2011) Vib Spectrosc 55:207–215
Hathaway BJ (1983) Copp Coord Chem Rev 52:87–169
Garribba E, Micera G (2006) J Chem Educ 83:1229–1232
Ray A, Mitra S, Khalaji AD, Atmani C, Cosquer N, Triki S, Clemente-Juan JM, Cardona-Serra S, Gómez-García CJ, Butcher RJ, Garribba E, Xu D (2010) Inorg Chim Acta 363:3580–3588
Nguyen DT, Le TH, Bui TTT (2013) Eur J Med Chem 60:199–207
Yousef TA, Abu-El-Reash GM, Al-Jahdali M, El-Rakhawy R (2014) Spectrochim Acta A 129:163–172
Firuzi O, Lacanna A, Petrucci R, Marrosu G, Saso L (2005) Biochim Biophys Acta 1721:174–184
El Anouar H, Gierschner J, Duroux J-L, Trouillas P (2012) Food Chem 131:79–89
Shubina VS, Shatalin YV (2013) J Appl Spectrosc 80:761–766
Brodowska K (2013) Biotechnol Food Sci 77(1):45–53
Popov LD, Gazieva GA, Kravchenko AN, Tkachenko YN, Askalepova OI, Levchenkov SI, Karpova TB, Tupolova YP, Kogan VA (2015) Russ J Gen Chem 85:92–96
Eshkourfu R, Cobeljić B, Vujcić M, Turel I, Pevec A, Sepcić K, Zec M, Radulović S, Srdić-Radić T, Mitić D, Andjelković K, Sladić D (2011) J Inorg Biochem 105:1196–1203
Shahabadi N, Mohammadi S (2012) Bioinorg Chem Appl. doi:10.1155/2012/571913
Acknowledgments
This work was financially support by Statute Funds No. I28/DzS/9184.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Brodowska, K., Sykuła, A., Garribba, E. et al. Naringenin Schiff base: antioxidant activity, acid–base profile, and interactions with DNA. Transition Met Chem 41, 179–189 (2016). https://doi.org/10.1007/s11243-015-0010-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11243-015-0010-7