Abstract
In this study, the complexity of a steady-state flow through porous media is revealed using confocal laser scanning microscopy (CLSM). Micro-particle image velocimetry (micro-PIV) is applied to construct movies of colloidal particles. The calculated velocity vector fields from images are further utilized to obtain laminar flow streamlines. Fluid flow through a single straight channel is used to confirm that quantitative CLSM measurements can be conducted. Next, the coupling between the flow in a channel and the movement within an intersecting dead-end region is studied. Quantitative CLSM measurements confirm the numerically determined coupling parameter from earlier work of the authors. The fluid flow complexity is demonstrated using a porous medium consisting of a regular grid of pores in contact with a flowing fluid channel. The porous media structure was further used as the simulation domain for numerical modeling. Both the simulation, based on solving Stokes equations, and the experimental data show presence of non-trivial streamline trajectories across the pore structures. In view of the results, we argue that the hydrodynamic mixing is a combination of non-trivial streamline routing and Brownian motion by pore-scale diffusion. The results provide insight into challenges in upscaling hydrodynamic dispersion from pore scale to representative elementary volume (REV) scale. Furthermore, the successful quantitative validation of CLSM-based data from a microfluidic model fed by an electrical syringe pump provided a valuable benchmark for qualitative validation of computer simulation results.
Graphic Abstract

Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Fluid flow through porous media is a multi-scale process that brings together fundamental laws of physics at the molecular level and empirical relations at macro scale level. We study an intermediate length scale (micrometer–millimeter) to explore difficulties associated with upscaling solute transport, i.e., hydrodynamic dispersion and mixing (Lowe and Frenkel 1996; Hunt et al. 2011; Boon et al. 2017; Svidrytski et al. 2019; Batany et al. 2019). The presented work is based on measurements and simulations of a sub-REV (Representative Elementary Volume) porous medium consisting of hundreds of pores.
We used microfluidic models to study laminar flow in porous media at the micrometer–millimeter length scale. A microfluidic model is a sheet of material containing a simple or complex network of small channels (see the review paper by Karadimitriou and Hassanizadeh 2012, for definitions of micromodels). A range of available manufacturing methods enable microfluidic technology to be used in a wide spectrum of applications. For example, there are a variety of sensors and analysis techniques, commonly with biomedical functionality, as reviewed by Sackmann et al. (2014), Ma et al. (2018) and Soitu et al. (2019). In addition, microfluidic techniques are emerging in heterogeneous catalysis (Xu et al. 2013; Zhang et al. 2019; Yue 2018) and advanced sensing technology (e.g., Hogan et al. 2017), among others. Microfluidic technology is part of the field of so-called lab-on-chip or microreactor devices. Crucial for the success of microfluidic technology is understanding the behavior of fluids in their channels (Squires and Quake 2005), for instance during mixing processes (Suh and Kang 2010). Fortunately, microscopical observations can be employed to investigate fluid mechanics in great detail due to the small dimensions of typical microfluidic channels. Matching experimental and computational efforts (see, e.g., Weishaupt et al. 2020, for single-phase flow, Kunz et al. 2016, for two-phase flow, and Zhang et al. 2013a, 2013b, for colloid transport during two-phase flow) deepens our understanding of fluid mechanics.
The challenge lies in extrapolating the results from microfluidic model studies to fluid mechanics at macro scale. Examples are the Beavers–Joseph interface condition for coupling free flow and porous media flow (Terzis et al. 2019), the motion of fluid–fluid interfaces of two immiscible fluids (Karadimitriou et al. 2014), transport of colloids during two-phase flow (Zhang et al. 2013a, b), and the movement of ganglia in porous media (Zarikos et al. 2018).
A variety of light microscopy techniques can be used to study fluid flow, in which one can employ various combinations of hardware (e.g., epi-fluorescence microscopy, spinning disk microscopy, confocal microscopy, super resolution microscopy, custom-made open microscopes), their corresponding acquisition strategies (volumetric imaging, optical slicing, deconvolution, astigmatism imaging, continuous/pulse imaging or scanning), and related digital post-processing steps (variety of track and trace algorithms). A full discussion on available techniques is beyond the scope of this work (Santiago et al. 1998; Park et al. 2004; Park and Kihm 2006; Cierpka and Kähler 2012; Klein et al. 2012; Kim et al. 2013; Kinoshita et al. 2006; Karadimitriou et al. 2012; Ichikawa et al. 2018, Guo et al. 2019).
In the present work, the microfluidic models were made from PDMS (polydimethylsiloxane) using soft lithography techniques (see, e.g., Karadimitriou et al. 2013). Typical cross-sectional dimensions of the channels are 100 × 100 µm2 or 200 × 100 µm2, while the length of the channels can be up to some tens of millimeters. The channels are fully saturated with demineralized water containing fluorescent colloidal particles. The density of these polystyrene colloids (1.05 gcm−3) is very similar to the density of water (0.997 gcm−3), and Brownian motion is negligible because of their diameter of 750 nm, which allows the colloids to act as an indicator for the movement of water.
A confocal laser scanning microscope (CLSM) is used to measure the velocity of the flow throughout the volume of the microfluidic channels (Park et al. 2004; Kinoshita et al. 2006; Silva et al. 2008). Truly 3D flow velocity measurements require observing fluid flow in all three directions, which is done by, for example, astigmatism imaging (Ichikawa et al. 2018). 3D flow velocity measurements are also known as 3D3C, which stands for three dimensions, three velocity components. Our CLSM work produces stacks of 2D vector fields, effectively being a 3D2C measurement. The velocity component in the third direction can be neglected when assuming that the surfaces of the microfluidic model are smooth, and the flow is in a laminar flow regime. In addition, the system must be stationary (i.e., steady-state flow). The stationary condition requirement allows sequential recording of multiple planes which, when stacked together, form the complete 3D2C measurement of a particular area in the microfluidic model. Moreover, multiple 3D2C measurement stacks of neighboring areas can be combined into a large volume of interest when stationary condition exists.
The fluorescence dye in the colloids is excited by the laser light from the confocal microscope and captured by the microscope lens system. The laser generates fluorescence throughout the entire thickness (z-axis) of the microfluidic model at a given lateral position (x, y). A pinhole in the optical path ensures that excited fluorescence only from a well-defined plane perpendicular to the z-axis reaches the detector. Effectively blocking the so-called “out-of-plane” light allows to record optical slices from a volume. As a result, observations can be made from a single point in 3D space with sub-micron resolutions in lateral directions and down to about a micron along the optical axis in our setup. A sample is typically imaged in 3D by using a motorized stage for the z-movement and moving mirrors to scan the laser laterally across the sample. The motorized stage also enables additional lateral movements.
Following the acquisition, a dedicated MATLAB-based software package PIVlab (Thielicke and Stamhuis 2014, Thielicke 2018) is used to analyze the collected data. PIVlab compares sets of consecutive images and determines the displacements of all particles visible in the images. The analysis results in 2D vector fields. Stacking of the vector fields results in the 3D2C measurement, the flow velocity throughout the volume of the microfluidic channels.
We compare the 3D2C observations with computer simulation results for practical reasons. Details of 3D2C measurements from specific geometries can be readily analyzed. However, too large structures become impractical from both an acquisition point of view and data post-processing.
What follows in this paper is a description of two different microfluidic models and for what purpose these microfluidic models have been used. Thereafter, the terms “streamline routing” and “hydrodynamic dispersion” are explained in the context of this work, as both terms are used throughout the rest of the paper.
1.1 Microfluidic Model Designs
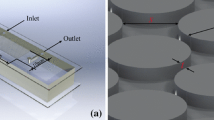
Two different microfluidic models have been designed. The first model consists of a horizontal channel, intersected by two narrower secondary channels perpendicular to it (see Fig. 1a). The depth of all channels is 0.15 mm. The two side channels have two different widths (I = 0.25 mm; II = 0.14 mm). This first microfluidic model is referred to as the T-junction model. The second microfluidic model consists of a large horizontal channel called the “free flow channel” which is bordering a network of channels called the porous region. The latter is created by square pillars positioned equidistantly (see Fig. 1b). The second model is referred to as the porous media coupling model (Terzis et al. 2019), as the design is used to study the coupling between the free flow channel and flow in the porous region.
The design of microfluidic models and the experimental setup. a The T-junction design consists of a free flow channel (yz-plane is 0.50 × 0.15 mm2), which can be fed from either side. The widths of the side channels are 0.25 mm (I) and 0.14 mm (II). b The porous media coupling design also has a free flow channel (I) which can be fed from both sides. The cross-sectional surface (yz-plane) is 2.0 x 0.2 mm2 and the opening to the porous region (II) is 38.64 mm long. The porous region consists of 80 × 20 uniformly spaced square pillars (0.24 × 0.24 mm) with pore spaces of 0.24 mm width (both horizontal and vertical). The height of the pillars is 0.20 mm. The triangular shaped region (III) is free from pillars and facilitates the saturation of the porous region through the auxiliary port. c A custom-made sample holder (yellow) supports the PDMS model on the piezo-driven stage of the confocal microscope
The T-junction model is used to study the coupling between free flow and an intersecting side channel, while the porous media coupling model is used for a benchmark test and to examine complex flow patterns. In the next section, we describe three different experiments that we have performed with our microfluidic setups.
1.1.1 Benchmark Test
In this study, we first quantitatively test the applicability of our CLSM setup for 3D2C measurements in complex porous media. As a benchmark test, we determine the flow through a single channel and compare the mean flow rate with the rate provided by the syringe pump. The example is also used to demonstrate a filtering technique that is developed in this study.
1.1.2 The Slip Velocity Across a T-Junction
In a second experiment, a full 3D2C measurement is performed for a free flow channel with intersecting side channels. Both the free flow channel and the intersecting side channels are fully saturated with water. The side channels are closed at the end, representing a dead-end region. Of interest is the coupling between the flow in the free flow channel and the movement within the dead-end region. Laminar flow through a straight tube exhibits a no-slip boundary (zero velocity of fluid) at the channel wall. The no-slip boundary is no longer valid when the side channel meets the porous structure, as the slip velocity becomes greater than zero along the plane between the free flow channel and porous media region. As a result, a momentum transfer occurs across the interface. The ability to measure the velocity along the interface region is particularly important for computer simulations, where the slip length serves as an input parameter. Earlier work by the authors has shown that a coupling can be made between a Navier–Stokes-type simulation of the free flow channel and a pore-network model-type simulation of intersecting side channels (Weishaupt et al. 2020). The pore-network-type modeling enables simulating large complex pore structures at very little computational cost. An accurate coupling of momentum transfer between the free flow channel and the porous media is crucial. It is shown that the coupling can be made through a slip coefficient βthroat, corresponding to the inverse value of the well-established concept of the Navier slip length applied to a single pore cavity. Previously, we determined the slip coefficient from simulations. In the current work, we compare the simulation data quantitatively with CLSM-based 3D2C measurements.
1.1.3 Complex Flow Patterns
The third experiment is set up to study complex flow patterns. The porous media coupling model is fully saturated with water before the auxiliary port is closed. A steady-state flow is established through the free flow channel. As a result, a flow is induced across the porous media which has been described by the Beavers–Joseph law (Terzis et al. 2019). CLSM-based 3D2C measurements are performed across parts of the porous region and compared to computer simulation.
1.2 Streamline Routing
Both confocal-based 3D2C measurements and computer simulations produce velocity vector maps. Therefore, it becomes possible to calculate streamlines by tracing the lines tangent to the velocity vectors. Streamlines do not cross under laminar flow conditions and represent a deterministic approach toward predicting solute pathways across porous media. Streamlines coming close together from different pores remain separated as if they are kept apart by an 'imaginary boundary' or barrier (Mehmani et al. 2014). A prerequisite for predicting solute transport is the ability to predict the routing of the pathways across complex pore systems, also known as streamline routing. Despite numerous attempts, streamline routing has not been resolved analytically for complex pore structures (Hull and Koslow 1986; Bruderer and Bernabé 2001; Mehmani and Tchelepi 2017), and it remains to be seen whether or not an analytical prediction is feasible. We reiterate the challenge by demonstrating the complexity of streamline routing through a very regular structure. In the next paragraph, we use the concept of streamlines to define hydrodynamic dispersion.
1.3 Hydrodynamic Dispersion
The pathway and the velocities along the pathway of a small packet of fluid in a stationary flow field can be fully described by a streamline. Under high Peclet number regime (where diffusion is negligible), the time of arrival of a solute following a streamline can be calculated for a given position along the streamline. The pressure-driven laminar flow of an incompressible Newtonian fluid in a straight pore (described by Hagen–Poiseuille flow) has a parabolic flow velocity profile (De Anna et al. 2017). Understandably, solutes travelling along streamlines in the center of the pore will arrive earlier at the end of the pore in comparison with solutes travelling along streamlines closer to the pore walls. Such difference in time of arrival is called longitudinal dispersion. Longitudinal dispersion in combination with diffusion (Brownian motion) in a straight tube has been solved analytically and is referred to as Taylor-Aris dispersion (Aris 1956; Raoof and Hassanizadehm 2010).
After discussing a single pore, we consider a porous medium with a large number of pores with varying pore throats. All pores have a mean and maximum velocity, which correspond to the specific pore throat geometry and the surrounding pores (e.g., De Anna et al. 2017). Again, a solute released simultaneously from one side of a porous media will not arrive simultaneously on the other side. The variation in time-of-arrival is a combination of longitudinal and so-called transversal dispersion, often lumped into “mechanical dispersion,” (e.g., Raoof and Hassanizadeh 2013; Svidrytski et al. 2019).
Mechanical dispersion directly follows from streamline routing at the pore-scale level. As discussed previously, streamline routing in complex pore structures has not been analytically resolved. Consequently, mechanical dispersion is likely to be equally difficult to be analytically resolved. Moreover, the Brownian motion of solutes must be considered at low flow velocities (i.e., small Peclet numbers). Consequently, a streamline with a high concentration of solute will show considerable diffusion of solute into neighboring streamlines with lower concentrations. Varying concentrations between neighboring streamlines originate from the variations in velocities of neighboring streamlines. The combined mechanical dispersion and diffusion is referred to as hydrodynamical dispersion. Visualizations of hydrodynamic dispersion with an increasing ratio between convection and diffusion can be found in, e.g., Martys (1994) and Icardi et al. (2014). Although hydrodynamical dispersion can be clearly defined qualitatively, there is no clear consensus regarding its implementation in mathematical frameworks (e.g., Lowe and Frenkel 1996; Hunt et al. 2011; Peters et al. 2019). Diffusion also invalidates the deterministic approach toward a description of individual particles or solutes being solely associated to streamlines, as neither particles nor solutes “stick” to their pathways. At the end of this paper, we will reflect on hydrodynamic dispersion, based on our observations.
2 Materials and Methods
2.1 Manufacturing a Microfluidic Model
The microfluidic models are made from PDMS (polydimethylsiloxane). The manufacturing process of PDMS microfluidic models by soft lithography techniques is described extensively elsewhere (Terzis et al. 2019 and references therein). A mask with the design of the microfluidic model is placed in between an UV source and a silicon wafer covered with a photoresist (SU-8) layer. Areas of the photoresist that are exposed to UV light harden. Subsequently, the remaining photoresist is washed off, leaving the silicon wafer with hardened photoresist features standing out, forming an inverted microfluidic model. The wafer is placed in a petri dish and covered with a layer of approximately 3–5-mm-thick PDMS (a ratio of 1:10 is used for the curing agent and polymer respectively) (Sylgard 184 Silicone Elastomer Kit). A second petri dish is filled with another layer of 3–5 mm of PDMS in order to make a flat slab for covering the main slab with the features. Subsequently, the petri dishes are placed in a vacuum chamber for several hours to ensure that all air bubbles leave the PDMS. Finally, both petri dishes are placed in an oven (60 °C) for at least 2 hours to cure and harden the PDMS.
Once the PDMS is cured, the wafer covered by the PDMS is taken out of the petri dish. The wafer is removed with great caution in order to not damage any PDMS micro features. Small holes are punched into the flat slab with the features, serving as inlet/outlets. A piece of PDMS with similar dimensions is cut from the flat PDMS slab. The two pieces of PDMS are fused together after creating dangling bonds at the surface by means of electrostatic discharging (BD-20A High Frequency Generator, ETP, Chicago, USA). The combined PDMS slabs are placed in the oven for at least 2 h before performing the experiments.
2.2 SEM Imaging of the Microfluidic Model
The exact dimensions of the channels in the microfluidic model have been determined by scanning electron microscopy (SEM) (Helios Nanolab UC-G3, Thermo Fisher Scientific, Eindhoven, the Netherlands). Prior to SEM imaging, a platinum sputter coating (Cressington HQ280, Cressington Scientific Instruments, Watford, UK) of ~ 6 nm is applied to the entire surface of the PDMS slab with the features. Such a thin metal layer ensures sufficient electrical conductivity at the surface of the PDMS to dissipate the charge from the electron beam. Images are recorded at 2 kV, 50 pA, using the Everhart–Thornley detector (ETD) in secondary electron (SE) mode.
Two samples are investigated: (1) The PDMS slab that came directly from the wafer and (2) two slabs of PDMS fused together. The latter is mounted on a pre-tilted holder to ensure a parallel view along the surface of the microfluidic model. All length measurements are corrected for stage tilt angles when applicable.
2.3 Microfluidic Setup
Fluorescent colloidal particles (Fluoresbrites YG Microspheres, Polysciences, Warrington (PA), USA) are added to the deionized water to be able to visualize the fluid motion. The polystyrene particles with a diameter of 750 nm and a density of 1.05 g/cm3 are assumed to move along streamlines. Steady-state fluid flows are generated using an electrical syringe pump (Pump 11, Pico Plus Elite, Harvard Apparatus, Holliston (MA), USA).
Full saturation of the microfluidic models must be ensured prior to an experiment. In the case of the T-junction model, slow infusion proves to be sufficient. Once full saturation is achieved, including the side channels, a plug is used to close the auxiliary ports of the side channel and a steady-state flow is established.
Achieving the full saturation of the coupling model is more challenging because of the hydrophobic nature of the PDMS. Therefore, a water–ethanol mixture is used to fully saturate the model through its auxiliary port. A home-made manual syringe pump is made by holding a syringe in place using a clamp and then operating the syringe by a pin with a fine screw thread that pushes against the syringe. Turning the pin manually, forcing the fluid out of the syringe, is found to be much easier controllable than pushing against the syringe by hand directly.
When the model is fully saturated, the auxiliary port (see Fig. 1b) is closed with a plug. Next, a constant flow of deionized water with fluorescent particles is initiated through the open channel via the inlet port. Care must be taken not to introduce air bubbles back into the microfluidic system. The entire model is flushed for at least 30 min at the rate of 1 ml/h to flush out the ethanol (Terzis et al. 2019).
2.4 Confocal Acquisition
Fluid flow is observed with a confocal microscope (Nikon A1R, Nikon Instruments, Amsterdam, the Netherlands) with the 488 nm laser. The utilized objective lens (CFI S Plan Fluor ELWD 20XC, magnification 20x, N.A. 0.45) results in 512x512 pixel images with a lateral (XY) scan resolution of 1.27 µm/pixel (covering a horizontal field of view of ~ 0.65 mm). A pinhole diameter is selected for different experiments in the range of 0.8–2 Airy Units. Resonant scanning combined with a serpentine scanning strategy results in a frame rate of 30 frames per second. One fluorescent channel and the camera image are recorded. The microscope is equipped with a piezo-driven stage to control the movements in all three directions.
Most observations require a larger field of view than the single 0.65 × 0.65 mm2 frame. In addition, each frame position consists of multiple image series, recorded at different z-positions. Therefore, Nikon’s JOBS software is used to program the microscope. JOBS allows for setting up an area, either a part of the microfluidic model or the entire microfluidic model. The total area is divided over n x m position matrix. A z-stack is defined per position and a short movie is recorded for every z-plane.
A typical movie consists of 50 to 75 frames. The step size along the z-axis is 100 µm and the number of steps is varied between 10 and 25 steps, depending on the depth of the channel. The range along the z-axis usually exceeds the thickness of the channel to ensure the entire channel is captured. As will be discussed below, the data can be digitally cropped at a later stage to exclude redundant volumes. Depending on the number of positions, the number of z-planes, and the length of each movie, total measurement times vary between 20 min and 8 h.
Prior to the acquisition, the focus distance is determined manually across the microfluidic model, as a part of the JOBS routine. The stage moves to a number of predetermined positions. Thereafter, the focus distance is manually selected and saved to that position. The focus distance in between the manually determined positions is based on interpolation.
2.5 Particle Image Velocimetry
The Nikon confocal microscope produces a large number of ND2 type files (one file per movie). An in-house developed MATLAB code is used to extract all fluorescent images and a single-camera image from a specific ND2-file into a target directory. The code utilizes software provided by Bio-Formats (Linkert et al. 2010). Subsequently, a binary image (mask) is created to distinguish the pore space from the solid phase.
A second MATLAB code is used to perform time-resolved digital particle image velocimetry analysis. An in-house developed MATLAB code loads the data into the PIVlab code (Thielicke and Stamhuis 2014; Thielicke 2018). Each movie is analyzed individually, which allows for parallel computing of multiple movies simultaneously.
PIVlab analyzes the movement of all particles between two consecutive frames and repeats this analysis for all sets of two frames of an entire movie. In the end, it produces two series of vector maps: one with horizontal velocity components (u) and one with vertical velocity components (v). The original confocal images are 512 × 512 pixels. The vector maps are 63 × 63 pixels with a spatial resolution of ~ 10 µm. For example, a movie consisting of 50 frames (25 image pairs) results in 25 horizontal and 25 vertical vector maps of 63 × 63 pixels. A median filter (kernel of 3 × 3) is applied to all individual vector maps to take out outliers. Thereafter, the 25 maps are combined by computing the median value for each (x, y)-coordinate, reducing the final result into two 63 × 63 pixel maps, one with the horizontal velocity component and one with the vertical velocity component. Note that longer movies also result in these single maps.
2.6 Stitching, Post-Processing, and 3D Analysis
An in-house MATLAB code is developed for the final processing step in the PIV analysis. First, all data (u- and v-vector maps, the camera images, and the masks) is sorted per z-plane. Second, the data is stitched together for each z-plane. All images have an overlap of approximately 10–15%. Therefore, alignment of camera images is based on minimizing the intensity (gray level) difference between overlapping areas. The camera image alignment coordinates are subsequently used to align and stitch the velocity matrices from the PIV analysis as well. As a final result, two large 3D matrices are filled with the u- and v-vector maps containing all raw data obtained from the experimental observations.
The measured flow velocities from PIVlab in pixels per frame need to be compared to the flow rate in ml/h provided by the electrical syringe pump. With a pixel size of 1.27 µm and a time per frame of 1/30 s, the conversion factor from pixel per frame to mm/s unit is 1.27 × 30/1000 = 0.0381. The free flow channel of the porous media coupling model has dimensions of 2.0 × 0.2 mm. Hence, a flow rate of 0.2 ml/h results in an average flow velocity vav = 200 mm3/(3600 s x ((2.0 mm x 0.2 mm) = 0.138 mm/s. In that case, the maximum velocity vmax in the center of the free flow channel would be around 0.2 mm/s (vmax = 3/2 vav) for a rectangular channel.
The converted data can be directly imported into ParaView (version 5.5.2) (Ahrens et al. 2005; Ayachit, 2015) for visualization and analysis. Alternatively, some advanced filtering and alignment can be applied. With the z-stack oftentimes recorded over a greater range than the depth of the microfluidic model, some z-planes only contain noise. These false results must be filtered out of the data prior to computing, for example, a mean velocity. In addition, it is quite challenging to align the microfluidic model perfectly flat with respect to the microscope for several reasons. After curing, the PDMS remains flexible. As a consequence, large PDMS models suspended over the objective lens may be bending a small amount. Such curvature will become immediately visible in the final volumetric result. Even if the PDMS model is supported sufficiently to avoid bending, its manufacturing process has several stages where a small tilt could be introduced into the model. That may happen when the microfluidic PDMS model in the petri dish is placed into the oven. In what follows, a filtering and alignment routine will be explained to overcome these issues.
All pore walls defining a rectangular channel are considered as no-slip boundaries (i.e., with zero velocity of the fluid). The high aspect ratio of the channel (ratio of width to its depth) produces a parabolic flow profile along the z-axis (depth) (Flekkoy et al. 1995). Although the parabolic flow profile can be recognized in the raw data, fitting is hampered because measurements from z-planes below and above the microfluidic channel produce some nonzero values (i.e., noises) and zero-velocities. A simple parabolic fit through all the values along the z-axis should exclude these contributions. Therefore, first a Gaussian curve is fitted to the absolute values of the measured data. We take advantage of the Gaussian curve property of not descending to negative values for the velocities. As a result, zeros and negative values (from noise) are fitted by the tails of the Gaussian curve. Once fitted, the Gaussian curve is used to identify the position and width of the parabolic flow profile. The pore walls (velocity is zero) are determined from evaluating the Gaussian curve and thresholding the cumulative distribution function at 0.05 and 0.95. The data points within the range of the Gaussian curve are used to fit the parabolic flow profile. As a result, a parabolic fit is determined for every x, y positions.
An advantage of fitting the full data set with parabolic fitting parameters is the ability to compute the data with a smaller step size along the z-axis, which effectively is an advanced interpolation method. In addition, the fits can all be shifted to start at z = 0, effectively aligning the entire data set. Once filtered and aligned, the data is exported to ParaView. All procedures described above are carried out by an in-house developed MATLAB code.
2.7 Comparison with Simulation Results
The experimental measurements are compared to simulation results. The Stokes equations for incompressible steady-state laminar flow are solved using a finite volume approach, as described in Weishaupt et al. 2020. In short: The entire microfluidic model, comprising the free flow channel, the porous region, and the triangular domain are meshed in 3D using uniform hexahedral, axis-aligned cells such that 20 cells per throat width in all directions are assigned. Grid convergence is assured and the open-source CFD tool OpenFOAM (Jasak 2009) is used to obtain the stationary flow field. A fixed gauge pressure of 1e-3 Pa is set at inlet of the free flow channel while a pressure of zero Pascal is set at the outlet. Since only creeping flow conditions (Re < 1 in the free flow channel) are considered in this work, the exact value of the boundary conditions is of secondary importance since the flow field can be scaled linearly for comparison with the experimental data.
2.8 New Image Filter for Para View
Most of the visualization in ParaView is performed with standard methods available within the software. Additionally, we have developed a custom Python plug-in. This specific filter allows visualization of the surface of the ‘imaginary boundary’ (Sect. 1.3) between sets of streamlines in 3D with a stream surface (Helman and Hesselink 1990). The surface is constructed in two steps: First, streamlines are seeded on a line orthogonal to the flow direction of the streamline sets. A triangular mesh is generated by computing a triangulation between points along adjacent streamlines to represent the surface. The streamline computation utilizes the “Stream Tracer” filter, available in the Visualization Toolkit (VTK) (Schroeder et al. 2006). Seed points are placed equidistantly along a source line with a parametrizable resolution. A streamline is generated at each position by integrating the flow field in forward and backward directions until a maximum streamline length is reached. The integration is computed using fourth-order Runge–Kutta and a fixed step size. In the second step, the stream surface is generated by inserting triangles between adjacent streamlines. To this end, the points corresponding to the individual streamline integration steps are connected by a triangular strip. A number of properties of the stream surface can be adjusted: The resolution, the maximum streamline length, the start and end points of the seed line and the step size of the seed line.
3 Results
3.1 The Microfluidic Model
Results from the SEM investigation are shown in Fig. 2. The soft lithography technique delivers PDMS microfluidic models with pore dimensions deviating only a few micrometers from the original design. Noticeable is the slight tapering of the vertical walls of both the pillars and the side wall of the free flow channel. Another feature is a small undercut of the pillars (see Fig. 2a). Finally, all vertical walls exhibit strong striations (i.e., roughness with an appearance of curtains), The amplitude of the roughness is of the order of a few micrometers. The sealing of the flat slab of PDMS on top of the pillars appears to be very tight (Fig. 2b), ruling out fluid movement in between the pillar surface and the PDMS surface.
SEM images of the microfluidic models. a A bird’s eye view of the microfluidic model without the PDMS top-layer cover, showing the regular pattern of pillars. b The cross-sectional view of a fully developed microfluidic model shows the tight fixation of the flat layer of PDMS on top of the pillars and the undercut of the pillars at their base
3.2 The 3D2C Analysis Procedure
The workflow of the 3D2C measurement is summarized in Figs. 3, 4 and 5 for a single z-stack. Figure 3a shows a typical image from a movie. Individual particles are traced in time (i.e., from one image to another) (Fig. 3b), ultimately producing a vector map (Fig. 3c). A single z-stack consists of a stack of movies, which are separately processed into corresponding vector maps. Figure 4 shows a full z-stack. The horizontal plane (xy) shows both the velocity magnitude and the direction. A vertical cross section visualizes the 3D2C. Despite the limited number of z-planes, a clear parabolic flow profile is recognized in both the horizontal and vertical direction, as expected for the rectangular pore shape in between the pillars.
PIV analysis. a a single frame of a movie. b a small spatial selection of consecutive frames from the movie. Vertical dotted lines are added as guide to visualize the movement of the individual colloidal particles. c the analysis of the entire movie by PIVlab results in a separate horizontal (u) and vertical (v) vector fields, which can be combined into a 2D vector filed
The 3D velocity profile. The bounding box indicates the volume of the data set (i.e., 0.65 × 0.65 × 0.25 mm). A single z-plane is shown, with both the velocity magnitude as well as velocity vectors. An xy-plane depicts the 3D2C velocity profile which corresponds to the shape of a rectangular channel
The velocity profile along the z-axis can be evaluated for all (x, y)-coordinates upon combining the PIV results. A Gaussian fit (green) is used to locate the peak and width of the measured data (yellow spheres), as it was found that a direct parabolic fit was susceptible to noise. Subsequently, a parabolic curve (blue) is fitted. The example shown comes from the center of the free flow channel of the porous media coupling model (Fig. 1b)
An example of the filtering and alignment procedure is shown in Fig. 5. Fitting a parabolic equation to the raw data would overestimate the width of the profile, because of the first point to the left. The Gaussian fit is much less susceptible to one or multiple data points away from the actual flow profile. The exclusion of data points based on the Gaussian fit results in an accurate parabolic fit.
3.3 Quantitative Comparison of Input and Output Flow
As a benchmark test, the inflow and outflow rates of flow through the porous media coupling model (Fig. 1b) have been determined by CLSM observations and compared quantitatively to the flow rate from the electrical syringe pump. Sections of the free flow channel in between the in/out ports and the porous media region are analyzed for three different flow rates: 0.1 ml/h, 0.2 ml/h, and 0.4 ml/h.
The width of the free flow channel exceeds the field of view of a single frame. Therefore, z-stacks (30 slices, 10-µm step size) at four consecutive positions were recorded and subsequently stitched together (Fig. 6a). The PIVlab analysis conditions were: 75 frames per movie, 30 frames per second, 512 × 512 pixels @1.27 µm per pixels, vector map is 63 × 63 pixels @ 10.32 µm per pixel. The resulting flow profiles are shown in Fig. 6b for a number of cross sections of the yz-plane. The yz-plane is perpendicular to the original acquisition plane (xy-plane). A single cross section is shown in Fig. 6c, clearly showing noise and local shifts along the z-axis. The shifts are likely to be caused by a shift in focus distance between different z-stacks and a non-horizontal positioning of the microfluidic model in the microscope. Filtering and aligning the 3D2C data by the Gaussian and parabolic fitting routine significantly improved the outcome, as shown in Fig. 6d. The noise is largely gone, and the parabolic profiles are all aligned at z = 0. Finally, the 3D2C data sets for three different flow rates are shown (Fig. 6e).
3D2C measurement. a the camera images from four z-stacks are stitched. The arrow indicates the position of flow direction and the scale bar represents 0.125 mm. b the raw PIV data is stacked together into a 3D reconstruction. Multiple cross sections (yz-plane) along the x-axis are shown, perpendicular to the flow direction. c an individual cross section (yz-plane) of the raw PIV data shows both misalignment and the generated noise. d the same cross-sectional areas as in c) after filtering and alignment, resulting in a shift toward y = 0. Finally, the aligned and filtered 3D2C results of the three different flow rates can be observed in e bird’s eye view and f side view (xz-plane)
A quantitative comparison is shown in Table 1. The mean flow velocity (mm/s) was computed from all horizontal (u-component) vectors with a value larger than zero. Note that in this particular case, a mean value can be computed from the entire volume, as a straight channel is measured with flow in a single direction. In case of more complicated pore structures, this is no longer valid.
It was found that the raw 3D2C measurements deviate from the expected values. This is largely attributed to the noise in the data. The result from the filtered data (‘parabolic fitting’) matches the expected values very accurately. As an indication of quantitative reproducibility, the comparison between the input- and output channels is reported as well, demonstrating the consistency of the method.
3.4 The Slip Coefficient βslip
A camera image of the microfluidic model containing the T-junctions (Fig. 1a) is shown in Fig. 7a and the corresponding velocity magnitude map is shown in Fig. 7b. The data set is obtained from two consecutive z-stacks of 25 steps (10 µm step size, 1.27 µm/pixel, 30 frames per second, 100 frames per movie, PIV analysis resolution: 10.32 µm/pixel). The dashed line I indicates the position of the cross-sectional plane (yz) shown in Fig. 7c. It is observed that the free flow channel is not positioned perfectly horizontal, as indicated in the figure. A clear advantage of utilizing the 3D capability of the CLSM is that it allows analyzing the central plane of the microfluidic model irrespective of the accuracy of the positioning in the CLSM.
The T-junction experiment. a a camera image with the white arrow indicating the flow direction and the black arrow pointing at a presumably air pocket in the PDMS solid phase. It appeared not to affect the measurement. b the velocity magnitude map with annotations indicating the locations of the cross-sectional planes I and II. c the cross-sectional plane I shows a small inclination of ~ 1.7° between the microfluidic model and the imaging plane of the CLSM. The observation is important in order to locate the correct position for measuring βslip. d the 3D2C flow velocity profile entering and leaving the dead-end pore. e the cross-sectional plane II shown from the top (after correcting the inclination as described in c) with the y-component of the velocity magnitude map
The coupling (i.e., momentum transfer) between a free flow channel and an intersecting dead-end pore can be parametrized by a slip coefficient βslip (Weishaupt et al. 2020) which is the inverse of the Navier slip length for a single cavity:
here <> is a surface average along a line across the center of the side channel in plane II (Fig. 7b), along which the velocity gradients ∂vx/∂y and ∂vy/∂x, and the mean slip velocity (vx) are determined, delineating the opening of the side channel. The analysis is performed in Paraview: The gradients are determined (Paraview module GradientOfUnstructuredDataSet), followed by the calculator set up to compute the sum of ∂vx/∂y (Gradient 1) and ∂vy/∂x (Gradient 3). The input values (numerator, denominator) for Eq. (1) come from the module DescriptiveStatistics applied to the previously defined line (module PlotOverLine).
The slip coefficient βslip has been determined for both auxiliary ports of the T-junction microfluidic model (Fig. 1a). The results are listed in Table 2. The same geometry has been simulated. The initial simulation result (not shown) was 16–20% off in comparison with the 3D2C measurement. A closer inspection of the geometry showed that the PDMS microfluidic model has rounded corners whereas the simulated model had sharp corners. Including the rounded corners in the simulated geometry resulted in an acceptable quantitative agreement between simulations and measurements (Table 2). The remaining deviation is attributed to the roughness of the PDMS.
With this, the quantitative comparison between experimental input, 3D2C measurement results, and simulations is demonstrated. Next, we observe the complexity of fluid flow through a porous structure.
3.5 Streamline Routing Through Porous Structures
During fluid flow across the entire microfluidic model, streamline routing is investigated in a small part of the porous structure of the porous media coupling model (Fig. 1b). The data set consists of 2 × 2 z-stacks with 25 steps of 12.5 µm each. Each movie consist of 75 frames with the following conditions: 512 × 512 pixels, 1.27 µm/pixel, 30 frames per second. The experimental data is displayed in Fig. 8. The area of interest is located away from the center of the porous media (i.e., Figure 9II), resulting in a somewhat diagonal flow across the regular pore structure. Streamlines are drawn in the center plane (z-axis) of the analyzed volume with the red color depicting a high flow velocity. It is obvious that solutes or particles travelling along a single streamline will experience both accelerations and decelerations. The 3D2C flow profiles indicate that a parabolic profile is developed very quickly in between the solid blocks before splitting up into the next intersection.
Experimentally-obtained streamlines and velocity profiles. The movement of fluid across complex pore structures has been analyzed by CLSM. The off-centered position (i.e., Figure 9II) results in a somewhat diagonal flow. The 3D2C flow profiles indicate the rapid development of a fully developed flow profile in between the solid blocks, with color coding indicating the flow velocity. Cross sections (I) and (II) show the time of arrival distribution across individual pores
Simulation-based streamlines represented using ParaView. The entire geometry (Fig. 1b) has been simulated, but streamlines are initiated from the middle vertical plane to the right, as indicated by the arrows. The color coding indicates the time integral, i.e., the required time to travel along a specific streamline up until a particular position
A comparison is made with simulation results (Figs. 9 and 10). The same diagonal pattern for the streamlines is observed in the inserts (Fig. 9). Immediately clear is the complexity of routes from the streamlines around the solid blocks, despite the regular geometry. The position of individual streamlines strongly alternates between the center of the pore and in the vicinity of a pore wall, with a corresponding high and low velocity, respectively.
a The imaginary boundaries between streamlines based on a 3D numerical simulation of the microfluidic model (Fig. 9). a Bird’s eye view perspective. b Top view. c Front view of the plane indicated in a)
Figure 9 is based on a 3D simulation. The behavior of streamlines in 3D is investigated in Fig. 10. Instead of the streamlines themselves, the ‘imaginary barriers’ (Mehmani et al. 2014) between streamlines are shown, based on the principles that streamlines in stationary laminar flow cannot cross. The imaginary barriers are mostly perpendicular to the xy-plane because the flow is laminar and the top and bottom of the simulated microfluidic model is perfectly smooth. Some curvature is observed in the planes (Fig. 10c).
In effect, streamline routing is established in 3D based on the simulation data which is qualitatively supported by the experimental data. The next step is determining the travel time along individual streamlines. The color coding in Fig. 9 indicates the travel time from the start of a streamline to the point of interest on the streamline. The continuously varying distance between a streamline and a pore wall inevitably creates a delay or fast forward with respect to neighboring streamlines. The shift in arrival times is clearly visible in Fig. 9 as blue and red colored lines are fully mixed. The two cross sections shown in Fig. 8 (labeled I and II) show the same mechanism in 3D from the 3D2C data set.
Finally, Fig. 11 captures both the velocity distribution (2D plane) and the time-of-arrival (cross sections in third dimension) in a single image. The velocity distribution maintains a parabolic profile in the pores connecting two consecutive junctions. The time-of-arrival forms a progressively more complicated pattern. Similar to the ‘imaginary barriers’, the pattern observed in Fig. 9 is extended into the third dimension, resulting in parallel planes with a relative shift in time-of-arrival. The no-slip boundary condition slows down the streamline close to the top and bottom of the network, regardless of streamline routing.
A larger section of the simulated domain (Fig. 9). The velocity profile is shown in the (x,y)-plane (opaque) while the multiple cross sections ((y,z)-planes) indicate the time-of-arrival at those positions. The generated flow in this part of the model is slightly diagonal as indicated by the arrows in the left corner. The time-of-arrival is measured from the cross-sectional plane in the back. The distribution of time-of-arrival across an individual pore is more or less constant between the top and bottom of the model, except where the streamlines move close to the top or bottom, where the no-slip boundary condition slows down the streamline regardless of the streamline routing
4 Discussion
4.1 PDMS Micromodels
The use of PDMS is advantageous because of its ease of use. A number of PDMS properties must be taken into account in order to prevent unreliable results:
-
PDMS is known to swell when in contact with some solvents (Lg Nee et al. 2003; Dangla et al. 2010). The presented work utilized demineralized water which is not expected to cause any swelling.
-
Pressure-induced deformation of PDMS is known to occur, but it is unlikely to occur for the pressure conditions and pore dimensions of our study (Gervais et al. 2006, Raj et al. 2018).
-
The manufacturing process and the mounting of the PDMS microfluidic models in the microscope are likely causing misalignment to some degree, as pointed out in Fig. 7c. Sources for misalignments range from uneven plastic petri dishes, uneven placement of the petri dish in the oven during curing and applying uneven pressure across the microfluidic model during bonding. Even though the channel geometry remains unaffected by a small inclination during the manufacturing process, the ability to place the microfluidic model perfectly flat in the CLSM is reduced.
-
The most significant source for misalignments is likely to be the mounting of the microfluidic model in the CLSM in combination with the flexibility of the PDMS slabs. Large PDMS slabs (> 2 cm) in particular will bent toward the center. As a result, moving the stage along the microfluidic model requires a continuous adjustment of the focus distance. To prevent bending, a dedicated holder is used to support the PDMS slabs. As a last resort, a strong advantage for using a CLSM is the opportunity to make corrections on the digitally reconstructed volume, as shown in Fig. 7.
4.2 3D2C Data Handling
A practical consideration is data handling of the 3D2C measurement. Automation of the acquisition and post-processing is crucial, as both require several hours up to several days, depending on the number of z-stacks, the number of steps per z-stack and the number of frames per step. Data shown in the present work is obtained from data sets which are each typically 3–10 GB. Post-processing is performed on a 32-core workstation with 64 GB RAM enabling much larger data sets (300 GB) to be processed in semi-automated overnight and weekend runs. Larger datasets impose increasing demands on visualization capabilities as well.
4.3 Comparison of 3D2C Measurements and Simulations
We have presented both a quantitative and qualitative comparison between 3D2C measurements in microfluidic devices and computer simulations of the same geometry. A comparison between 3D2C measurements and simulated data that is very helpful, complimentary and non-trivial. Care must be taken to ensure that the simulated geometry actually corresponds to the microfluidic device. A design with sharp 90 degrees corners will in reality contain rounded corners. Not considering rounded corners led to a deviation up to 20% when measuring the slip coefficient βslip.
The extensive sequence of events during acquisition and post-processing of the experimental data makes it plausible that structural measurement errors are introduced. Hence, a quantitative validation is performed. Table 1 provides clear evidence of the reliability of the data acquisition and post-processing. Next, a quantitative comparison is made with computer simulations by measuring the slip coefficient βslip (Sect. 3.5). The flow conditions in the 3D2C measurements and the simulations are laminar. Consequently, velocities are linearly scalable, as inertia can be neglected. Hence, simulations do not need to simulate conditions exactly matching the experimental conditions. The scalability is also reflected in the agreement of the slip coefficient βslip between simulations and 3D2C measurements.
The ability to match the 3D2C measurements and the simulation results is also important in the next discussion about dispersion in porous media, as particular observations (e.g., complexity of streamline routing) can be found in experimental data and further investigated by simulations.
4.4 Dispersion in Porous Media
Quantifying hydrodynamic dispersion in porous media remains a scientific challenge. Only a limited number of analytical solutions are available (e.g., Aris 1956; Kreft and Zuber 1978), based on the advection diffusion equation (also known as the convection diffusion equation). However, the advection diffusion equation itself is considered doubtful regarding its applicability toward deriving a hydrodynamic dispersion coefficient (Hunt et al. 2011; Peters et al. 2019). Potential issues are the scale dependency of the hydrodynamic dispersion coefficient and the inability to fully describe breakthrough curves, for the latter in particular the lengthy tailing encountered in experimental and simulated data. Hydrodynamic dispersion is most often considered to be a Fickian diffusion process. Hence, any deviation from experimental or simulation data is called pre-asymptotic non-Fickian dispersion (Dentz et al. 2018; Puyguiraud et al. 2019 and references therein), or anomalous dispersion. The ‘pre-asymptotic’ term refers to the assumption that hydrodynamic dispersion can in fact be described by the advection diffusion equation at longer length- or time scales. That assumption is challenged when considering the hydrodynamic dispersion coefficient being scale dependent or not converging upon increasing the Peclet number (Lowe and Frenkel 1996). Experimental data also points toward nonlinear contributions to a hydrodynamic dispersion coefficient (Hunt et al. 2011; Boon et al. 2017; Svidrytski et al. 2019; Batany et al. 2019).
Hydrodynamic dispersion of solute in porous media is essentially the combined effect of streamline routing and Brownian motion. Even though the velocity profile in a single pore is parabolic (Figs. 8 and 11), the time-of-arrival profile within a single pore is much more complex (Figs. 8, 9, 10, and 11). It comes back to the earlier statement that streamline routing can only be derived from experimental observations (e.g., Morales et al. 2017; this work) or from numerical simulations. Notice that the work by Morales et al. utilized 68 µm diameter spheres, a size which effectively rules out diffusion. It does nicely fit in a very elegant line of work that studies Lagrangian (i.e., particle trajectories, so effectively streamlines) stochastics (Bijeljic et al. 2004; Dentz et al. 2016, 2018; Puyguiraud et al. 2019), and even including absorption/desorption (Miele et al. 2019). The work by Bijeljic stands out because of the use of pore-network models. A general feature of many pore-network models is the implicit assumption of full mixing at the nodes which is only valid at low flow rates (Mehmani et al. 2014; Sadeghi et al. 2020). Bijeljic accounts for mixing at the nodes by placing particles in pores through either an area-weighted distribution or pressure-weighted distribution. However, such strategy does not retain the history of individual streamlines. As such, it is not possible to obtain a time-of-arrival figure as in Figs. 8, 9, 10, and 11. The work by Dentz and co-workers is based on a continuous time random walk (CTRW) approach. It must be noted that again streamline routing is not explicitly accounted for. A strictly Lagrangian approach (Dentz et al. 2016) does include streamline routing, when the stochastic Lagrangian velocity distribution is recovered from actual measurements or simulations. Streamline routing is no longer included when the stochastic Lagrangian velocity distribution is derived from a stochastic description of the pore space (Dentz et al. 2018). Nevertheless, the CTRW approach appears to work well for colloidal particles (Morales et al. 2017; Miele et al. 2019). Proving the current form of CTRW to be suitable for molecular solutes would be the next step. Small concentrations of colloidal particles can diffuse freely across streamlines. In case of molecular solutions, concentration differences between neighboring streamlines dictate the net exchange of solute (Fickian diffusion) and thus strongly rely on the history of the interacting streamlines up to that point of interest. Figures 8, 9, 10, and 11 indicate that keeping track of the history of neighboring streamlines is not trivial. We must stress that our microfluidic models behave as two-dimensional models at low flow rates. A real three-dimensional pore structure would quite literally impose an extra dimension to the complexity. Therefore, we propose that future work will need to concentrate on establishing a predictive description of streamline routing. We foresee that it is likely to be a stochastic description, given the complexity of the pore space of porous media.
5 Conclusions
Confocal laser scanning microscopy allows for imaging fluid movement in microfluidic models. The presented method for reconstructing the volumetric flow profile (3D2C) is sufficiently accurate for quantitative analysis. Quantitative accuracy has been validated by comparing the flow velocity from a 3D2C measurement to the flow velocity provided by a syringe pump. Additional evidence of experimental accuracy is provided by measuring a slip coefficient βslip in a T-junction with a dead-end pore. It is noticed that the exact details of the geometry of the microfluidic model are crucial when it comes to using computer models. Our T-junction microfluidic models exhibit rounded corners at the junction. The comparison between experiments and computer simulations showed the necessity of including rounded corners in computer simulations.
The complexity of fluid movement in porous media is fully captured by CLSM, as shown in both the T-junction and the porous media coupling model. The latter clearly shows the ‘streamline routing’ effect in laminar flow. Suppose a high flow rate is combined with a relatively low diffusion coefficient (i.e., under high Peclet number). Such combination results in non-mixing behavior in pore bodies. Not only does it complicate any mathematical treatment of porous media flow, it also challenges the validity of pore-network models at large Peclet numbers because of unpredictable streamline routing. Most pore-network models implicitly assume full mixing in the pore bodies (i.e., nodes).
Another complexity is found in dispersion mechanisms, as the time for a solute or particle to travel along a specific streamline is highly dependent on the routing of that streamline. Consequently, diffusion between neighboring streamlines may occur back and forth at different points in time and space. Being able to properly take these complexities into account, will most likely result in an improved ability to predict solute transport across porous media.
Code availability
The in-house developed codes are available upon request.
References
Ahrens, J., Geveci, B., Law, C.: ParaView: An End-User Tool for Large Data Visualization. Elsevier, Visualization Handbook (2005)
Aris, R.: On the dispersion of a solute in a fluid flowing through a tube. Proc. Roy. Soc. A. 235, 67–77 (1956)
Ayachit, U.: The ParaView Guide: A Parallel Visualization Application. Kitware. (2015)
Batany, S., Peyneau, P.-E., Lassabatère, L., Béchet, B., Faure, P., Dangla, P.: Interplay between molecular diffusion and advection during solute transport in macroporous media. Vadose Zone J. 18, 180140 (2019)
Bijeljic, B., Muggeridge, A.H., Blunt, M.J.: Pore-scale modeling of longitudinal dispersion. Water Resour. Res. 40, 11501 (2004)
Boon, M., Bijeljic, B., Krevor, S.: Observations of the impact of rock heterogeneity on solute spreading and mixing. Water Resour. Res. 53, 4624–4642 (2017)
Bruderer, C., Bernabé, Y.: Network modeling of dispersion: transition from Taylor dispersion in homogeneous networks to mechanical dispersion in very heterogeneous ones. Water Resour. Res. 37, 897–908 (2001)
Cierpka, C., Kähler, C.J.: Particle imaging techniques for volumetric three-component (3D3C) velocity measurements in microfluidics. J. Vis. 15, 1–31 (2012)
Dangla, R., Gallaire, F., Baroud, C.N.: Microchannel deformations due to solvent-induced PDMS swelling. Lab Chip 10, 2972–2987 (2010)
De Anna, P., Quaife, B., Biros, G., Juanes, R.: Prediction of the low-velocity distribution from the pore structure in simple porous media. Phys. Rev. Fluids. 2, 124103 (2017)
Dentz, M., Kang, P.K., Comolli, A., Le Borgne, T., Lester, D.R.: Continuous time random walks for the evolution of Lagrangian velocities. Phys. Rev. Fluids 1, 074004 (2016)
Dentz, M., Icardi, M., Hidalgo, J.J.: Mechanisms of dispersion in a porous medium. J. Fluid Mech. 841, 851–882 (2018)
Flekkoy, E.G., Oxaal, U., Feder, J., Jøssang, T.: Hydrodynamic dispersion at stagnation points: simulations and experiments. Phys. Rev. E 52, 4952–4962 (1995)
Gervais, T., El-Ali, J., Günther, A., Jensen, K.F.: Flow-induced deformation of shallow microfluidic channels. Lab Chip 6, 500–507 (2006)
Guo, T., Ardekani, A.M., Vlachos, P.P.: Microscale, scanning defocusing volumetric particle-tracking velocimetry. Exp. Fluids 60, 89 (2019)
Helman, J.L., Hesselink, L.: Surface representations of two-and three-dimensional fluid flow topology. IEEE Proceed. First IEEE Conf. Vis. Vis. 90, 6–13 (1990)
Hogan, B.T., Dyakov, S.A., Brennan, L.J., Younesy, S., Perova, T.S., Gun’ko, Y.K., Craciun, M.F., Baldycheva, A.: Dynamic in situ sensing of fluid-dispersed 2D materials integrated on microfluidic Si chip. Sci. Rep. (2017). https://doi.org/10.1038/srep42120
Hull, L.C., Koslow, K.N.: Streamline routing through fracture junctions. Water Resour. Res. 22, 1731–1734 (1986)
Hunt, A.G., Skinner, T.E., Ewing, R.P., Ghanbarian-Alavijeh, B.: Dispersion of solutes in porous media. Eur. Phys. J. B 80, 411–432 (2011)
Icardi, M., Boccardo, G., Marchisio, D.L., Tosco, T., Sethi, R.: Pore-scale simulation of fluid flow and solute dispersion in three-dimensional porous media. Phys. Rev. E 90, 013032 (2014)
Ichikawa, Y., Yamamoto, K., Motosuke, M.: Three-dimensional flow velocity and wall shear stress distribution measurement on a micropillar-arrayed surface using astigmatism PTV to understand the influence of microstructures on the flow field. Microfluid. Nanofluid. 22, 73 (2018)
Jasak, H.: OpenFOAM: open source CFD in research and industry. Int. J. Nav. Arch. Ocean 1, 89–94 (2009)
Karadimitriou, N.K., Hassanizadeh, S.M.: A review of micromodels and their use in two-phase flow studies. Vadose Zone J (2012). https://doi.org/10.2136/vzj2011.0072
Karadimitriou, N.K., Joekar-Niassar, V., Hassanizadeh, S.M., Kleingeld, P.J., Pyrak-Nolte, L.J.: A novel deep reactive ion etched (DRIE) glass micro-model for two-phase flow experiments. Lab Chip 12, 3413–3418 (2012)
Karadimitriou, N.K., Musterd, M., Kleingeld, P.J., Kreutzer, M.T., Hassanizadeh, S.M., Joekar-Niasar, V.: On the fabrication of PDMS micromodels by rapid prototyping, and their use in two-phase flow studies. Water Resour. Res. 49, 2056–2067 (2013)
Karadimitriou, N.K., Hassanizadeh, S.M., Joekar-Niassar, V., Kleingeld, P.J.: Micromodel study of two-phase flow under transient conditions: quantifying effects of specific interfacial area. Water Resour. Res. 50, 8125–8140 (2014)
Kim, H., Westerweel, J., Elsinga, G.E.: Comparison of tomo-PIV and 3D-PTV for microfluidic flows. Meas. Sci. Technol. 24, 024007 (2013)
Kinoshita, H., Kaneda, S., Fujii, T., Oshima, M.: Three-dimensional measurement and visualization of internal flow of a moving droplet using confocal micro-PIV. Lab Chip 7, 338–346 (2006)
Klein, S.A., Moran, J.L., Frakes, D.H., Posner, J.D.: Three-dimensional three component particle velocimetry for microscale flows using volumetric scanning. Meas. Sci. Technol. 23, 085304 (2012)
Kreft, A., Zuber, A.: On the physical meaning of the dispersion equation and its solution for different initial and boundary conditions. Chem. Eng. Sci. 33, 1471–1480 (1978)
Kunz, P., Zarikos, I., Karadimitriou, N.K., Huber, M., Nieken, U., Hassanizadeh, S.M.: Study of multi-phase flow in porous media: comparison of SPH simulations with micro-model experiments. Trans. Porous Media 114, 1–20 (2016)
Lg Nee, J., Park, C., Whitesides, G.M.: Solvent compatibility of poly(dimethylsiloxane)-based microfluidic devices. Anal. Chem. 75, 6544–6554 (2003)
Linkert, M., Rueden, C.T., Allan, C., Burel, J.-M., Moore, W., Patterson, A., Loranger, B., Moore, J., Neves, C., MacDonald, D., Tarkowska, A., Sticco, C., Hill, E., Rossner, M., Eliceiri, K.W., Swedlow, J.R.: Metadata matters: access to image data in the real world. J. Cell Biol. 189, 777–782 (2010)
Lowe, C.P., Frenkel, D.: Do hydrodynamic dispersion coefficients exist? Phys. Rev. Lett. 77, 4552–4555 (1996)
Ma, Y.-H.V., Middleton, K., You, L., Sun, Y.: A review of microfluidic approaches for investigating cancer extravasation during metastasis. Microsyst. Nanoeng. (2018). https://doi.org/10.1038/micronano.2017.104
Martys, N.S.: Fractal growth in hydrodynamic dispersion through random porous media. Phys. Rev. E 50, 335–342 (1994)
Mehmani, Y., Oostrom, M., Balhoff, M.T.: A streamline splitting pore-network approach for computationally inexpensive and accurate simulation of transport in porous media. Water Resour. Res. 50, 2488–2517 (2014)
Mehmani, Y., Tchelepi, H.A.: Multiscale computation of pore-scale fluid dynamics: single-phase flow. J. Comput. Phys. 375, 1469–1487 (2017)
Miele, F., De Anna, P., Dentz, M.: Stochastic model for filtration by porous media. Phys. Rev. Fluids 4, 094101 (2019)
Morales, V.L., Dentz, M., Willmann, M., Holzner, M.: Stochastic dynamics of intermittent pore-scale particle motion in three-dimensional porous media: experiments and theory. Geophys. Res. Lett. 44, 9361–9371 (2017)
Park, J.S., Choi, C.K., Kihm, K.D.: Optically sliced micro-PIV using confocal laser scanning microscopy (CLSM). Exp. Fluids 37, 105–119 (2004)
Park, J.S., Kihm, K.D.: Three-dimensional micro-PTV using deconvolution microscopy. Exp. Fluids 40, 491–499 (2006)
Peters, A., Iden, S.C., Durner, W.: Local solute sinks and sources cause erroneous dispersion fluxes in transport simulations with the convection-dispersion equation. Vadose Zone J. 18, 190064 (2019)
Puyguiraud, A., Gouze, P., Dentz, M.: Stochastic dynamics of Lagrangian pore-scale velocities in three-dimensional porous media. Water Resour. Res. 55, 1196–1217 (2019)
Raj, A., Suthanthiraraj, P.P.A., Sen, A.K.: Pressure-driven flow through PDMS-based flexible microchannels and their applications in microfluidics. Microfluid. Nanofluid. 22, 128 (2018)
Raoof, A., Hassanizadehm, S.M.: Upscaling transport of adsorbing solutes in porous media. J. Porous Media 13, 395–408 (2010)
Raoof, A., Hassanizadeh, S.M.: Saturation-dependent solute dispersivity in porous media: pore-scale process. Water Resour. Res. 49, 1943–1951 (2013)
Sackmann, E.K., Fulton, A.L., Beebe, D.J.: The present and future role of microfluidics in biomedical research. Nature 507, 181–189 (2014)
Sadeghi, M.A., Agnaou, M., Barralet, J., Gostick, J.: Dispersion modeling in pore networks: a comparison of common pore-scale models and alternative approaches. J. Contam. Hydro. 228, 103578 (2020)
Santiago, J.G., Wereley, S.T., Meinhart, C.D., Beebe, D.J., Adrian, R.J.: A particle image velocimetry system for microfluidics. Exp. Fluids 25, 316 (1998). https://doi.org/10.1007/s003480050235
Schroeder, W., Martin, K., Lorensen, B. (2006) The Visualization Toolkit (4th ed.), Kitware
Silva, G., Leal, N., Semiao, V.: Micro-PIV and CFD characterization of flows in a microchannel: velocity profiles, surface roughness and Poiseulle numbers. Int. J. Heat Fluid Flow 29, 1211–1220 (2008)
Soitu, C., Feuerborn, A., Deroy, C., Castrejón-Pita, A.A., Cook, P.R., Walsh, E.J.: Raising fluid walls around living cells. Sci. Adv. 1, 1 (2019)
Squires, T.M., Quake, S.R.: Microfluidics: fluid physics at the nanoliter scale. Rev. Modern Phys. 77, 977–1024 (2005)
Suh, Y.K., Kang, S.: A review on mixing in microfluidics. Micromachines 1, 82–111 (2010)
Svidrytski, A., Hlushkou, D., Tallarek, U.: Relationship between bed heterogeneity, chord length distribution, and longitudinal dispersion in particulate beds. J. Chromatogr. A 1600, 167–173 (2019)
Terzis, A., Zarikos, I., Weishaupt, K., Yang, G., Chu, X., Helmig, R., Weigand, B.: Microscopic velocity field measurements inside a regular porous medium adjacent to a low Reynolds number channel flow. Phys. Fluids 31, 042001 (2019)
Thielicke, W., Stamhuis, E.J.: PIVlab—towards user-friendly, affordable and accurate digital particle image velocimetry in MATLAB. J. Open Res. Softw. (2014). https://doi.org/10.5334/jors.bl
Thielicke, W. (2018) PIVlab—particle image velocimetry (PIV) tool (https://www.mathworks.com/matlabcentral/fileexchange/27659-pivlab-particle-image-velocimetry-piv-tool). MATLAB Central File Exchange. Retrieved November 28, 2018
Weishaupt, K., Terzis, A., Zarikos, I., Yang, G., Flemisch, B., De Winter, D.A.M., Helmig, R. (2020) A hybrid-dimensional coupled pore-network/free-flow model including pore-scale slip and its application to a micromodel experiment. Submitted to Transport in Porous Media
Xu, B.-B., Zhang, Y.-L., Wei, S., Ding, H., Sun, H.-B.: On-Chip catalytic microreactors for modern catalysis research. ChemCatChem 5, 2091–2099 (2013)
Yue, J.: Multiphase flow processing in microreactors combined with heterogeneous catalysis for efficient and sustainable chemical synthesis. Catal. Today 308, 3–19 (2018)
Zarikos, I., Terzis, A., Hassanizadeh, S.M., Weigand, B.: Velocity distributions in trapped and mobilized non-wetting phase ganglia in porous media. Sci. Rep. 8, 13228 (2018)
Zhang, Q., Hassanizadeh, S.M., Karadimitriou, N.K., Raoof, A., Liu, B., Kleingeld, P.J., Imhof, A.: Retention and remobilization of colloids during steady-state and transient two-phase flow. Water Resour. Res. 49, 8005–8016 (2013a)
Zhang, Q., Karadimitriou, N.K., Hassanizadeh, S.M., Kleingeld, P.J., Imhof, A.: Study of colloids transport during two-phase flow using a novel polydimethylsilocane micro-model. J. Colloid Interface Sci. 401, 141–147 (2013b)
Zhang, J., Chen, J., Peng, S., Peng, S., Zhang, Z., Tong, Y., Miller, P.W., Yan, X.-P.: Emerging porous materials in confined spaces: from chromatography applications to flow chemistry. Chem. Soc. Rev. 48, 2566–2595 (2019)
Acknowledgements
We would like to acknowledge Ioannis Zarikos, Enno de Vries, Hamed Aslannejad, and Emma Verver (Nikon) for helping and discussing practical aspects of the work. The fist author is funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project Number 327154368–SFB 1313.
Funding
Deutsche Forschungsgemeinschaft (DFG, German Research Foundation)—Project Number 327154368–SFB 1313.
Author information
Authors and Affiliations
Contributions
MdW designed and performed the CLSM experiments, data processing and wrote the manuscript. KW designed and performed the computer simulations. SS and SF developed Paraview tools. KW, SS, SF, AR, SMH and RH made significant scientific contributions to data interpretation and were actively involved in preparing the manuscript.
Corresponding author
Ethics declarations
Conflicts of interests
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
de Winter, D.A.M., Weishaupt, K., Scheller, S. et al. The Complexity of Porous Media Flow Characterized in a Microfluidic Model Based on Confocal Laser Scanning Microscopy and Micro-PIV. Transp Porous Med 136, 343–367 (2021). https://doi.org/10.1007/s11242-020-01515-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11242-020-01515-9