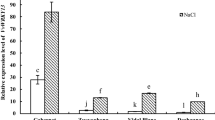
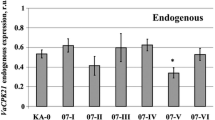
Abstract
Mitogen-activated protein kinase pathways are involved in plant resistance to a variety of adverse environmental processes, and their downstream component MAPKs play an important role in this process. However, the function of MAPKs in abiotic stresses is still far from being clear in grape (Vitis vinifera L.). Here, we isolated a novel group B MAPK gene (VvMAPK9) from grape, which is induced by different abiotic stresses such as salt, drought and high temperature (42 °C). Overexpressing VvMAPK9 in Arabidopsis thaliana significantly enhanced the tolerance to salt stress. Compared with wild type plants, the transgenic lines exhibited higher germination rate and longer root length as well better growth status under salt stress. In addition, overexpression of VvMAPK9 in grape callus also increased the salt stress tolerance and enhanced the callus’s ability to scavenge reactive oxygen species (ROS), which correlated with higher activity of ROS-related antioxidant enzymes. These results indicate that VvMAPK9 may positively regulate salt stress by regulating the antioxidative system.
Key message
Grape VvMAPK9 positively regulates salt tolerance in Arabidopsis and grape callus through regulating the antioxidative system








Similar content being viewed by others
Data availability
The amino acid sequences of Arabidopsis was downloaded from The Arabidopsis Information Resource (https://www.arabidopsis.org).
Code availability
Amino acid sequences of other plants MAPK genes were retrieved from GenBank (http://www.ncbi.gov/Genbank). Amino acid sequence alignments were done using the DNAMAN program (version 5.2.2). Analysis of the promoter sequence of VvMAPK9 were performed using PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/). The phylogenetic tree was constructed by the NJ (Neighbor-Joining) method using MEGA 4. Statistical significance was analyzed using Duncan’s multiple range tests with analysis of variance (ANOVA), and calculations were performed with SPSS Statistics.
References
Asai S, Ohta K, Yoshioka H (2008) MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20(5):1390–1406. https://doi.org/10.1105/tpc.107.055855
Bohnert HJ, Nelson DE, Jensen RG (1995) Adaptations to environmental stresses. Plant Cell 7(7):1099–1111. https://doi.org/10.1105/tpc.7.7.1099
Brader G, Djamei A, Teige M, Palva ET, Hirt H (2007) The MAP kinase kinase MKK2 affects disease resistance in Arabidopsis. Mol Plant Microbe Interact 20(5):589–596. https://doi.org/10.1094/MPMI-20-5-0589
Çakır B, Kılıçkaya O (2015) Mitogen-activated protein kinase cascades in Vitis vinifera. Front Plant Sci 6:556. https://doi.org/10.3389/fpls.2015.00556
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16(6):735–743. https://doi.org/10.1046/j.1365-313x.1998.00343.x
Danquah A, de Zelicourt A, Colcombet J, Hirt H (2014) The role of ABA and MAPK signaling pathways in plant abiotic stress responses. Biotechnol Adv 32(1):40–52. https://doi.org/10.1016/j.biotechadv.2013.09.006
Danquah A, de Zélicourt A, Boudsocq M, Neubauer J, Frei Dit Frey N, Leonhardt N, Pateyron S, Gwinner F, Tamby JP, Ortiz-Masia D, Marcote MJ, Hirt H, Colcombet J (2015) Identification and characterization of an ABA-activated MAP kinase cascade in Arabidopsis thaliana. Plant J 82(2):232–244. https://doi.org/10.1111/tpj.12808
Furuya T, Matsuoka D, Nanmori T (2014) Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Lett 588(11):2025–2030. https://doi.org/10.1016/j.febslet.2014.04.032
Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol Biochem 48(12):909–930. https://doi.org/10.1016/j.plaphy.2010.08.016
Gu L, Liu Y, Zong X, Liu L, Li DP, Li DQ (2010) Overexpression of maize mitogen-activated protein kinase gene, ZmSIMK1 in Arabidopsis increases tolerance to salt stress. Mol Biol Rep 37(8):4067–4073. https://doi.org/10.1007/s11033-010-0066-6
Hamel LP, Nicole MC, Sritubtim S, Morency MJ, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J, Martin G, Mundy J, Ohashi Y, Scheel D, Sheen J, Xing T, Zhang S, Seguin A, Ellis BE (2006) Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci 11(4):192–198. https://doi.org/10.1016/j.tplants.2006.02.007
Ichimura K, Mizoguchi T, Yoshida R, Yuasa T, Shinozaki K (2000) Various abiotic stresses rapidly activate Arabidopsis MAP kinases ATMPK4 and ATMPK6. Plant J 24(5):655–665. https://doi.org/10.1046/j.1365-313x.2000.00913.x
Jalmi SK, Sinha AK (2015) ROS mediated MAPK signaling in abiotic and biotic stress-striking similarities and differences. Front Plant Sci 6:769. https://doi.org/10.3389/fpls.2015.00769
Jonak C, Ligterink W, Hirt H (1999) MAP kinases in plant signal transduction. Cell Mol Life Sci 55(2):204–213. https://doi.org/10.1007/s000180050285
Kovtun Y, Chiu WL, Tena G, Sheen J (2000) Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA 97(6):2940–2945. https://doi.org/10.1073/pnas.97.6.2940
Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63(4):1593–1608. https://doi.org/10.1093/jxb/err460
Li Y, Cai H, Liu P, Wang C, Gao H, Wu C, Yan K, Zhang S, Huang J, Zheng C (2017) Arabidopsis MAPKKK18 positively regulates drought stress resistance via downstream MAPKK3. Biochem Biophys Res Commun 484(2):292–297. https://doi.org/10.1016/j.bbrc.2017.01.104
Liang Y, Chen Q, Liu Q, Zhang W, Ding R (2003) Exogenous silicon (Si) increases antioxidant enzyme activity and reduces lipid peroxidation in roots of salt-stressed barley (Hordeum vulgare L.). J Plant Physiol. 160(10):1157–1164. https://doi.org/10.1078/0176-1617-01065
Lu Y, Su W, Bao Y, Wang S, He F, Wang D, Yu X, Yin W, Liu C, Xia X (2020) Poplar PdPTP1 gene negatively regulates salt tolerance by affecting ion and ROS homeostasis in populus. Int J Mol Sci 21(3):1065. https://doi.org/10.3390/ijms21031065
MAPK Group (2002) Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci 7(7):301–308. https://doi.org/10.1016/s1360-1385(02)02302-6
Nakagami H, Pitzschke A, Hirt H (2005) Emerging MAP kinase pathways in plant stress signalling. Trends Plant Sci 10(7):339–346. https://doi.org/10.1016/j.tplants.2005.05.009
Pérez-Salamó I, Papdi C, Rigó G, Zsigmond L, Vilela B, Lumbreras V, Nagy I, Horváth B, Domoki M, Darula Z, Medzihradszky K, Bögre L, Koncz C, Szabados L (2014) The heat shock factor A4A confers salt tolerance and is regulated by oxidative stress and the mitogen-activated protein kinases MPK3 and MPK6. Plant Physiol 165(1):319–334. https://doi.org/10.1104/pp.114.237891
Qin F, Shinozaki K, Yamaguchi-Shinozaki K (2011) Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol 52(9):1569–1582. https://doi.org/10.1093/pcp/pcr106
Shi J, Zhang L, An H, Wu C, Guo X (2011) GhMPK16, a novel stress-responsive group D MAPK gene from cotton, is involved in disease resistance and drought sensitivity. BMC Mol Biol 12:22. https://doi.org/10.1186/1471-2199-12-22
Sun W, Chen H, Wang J, Sun HW, Yang SK, Sang YL, Lu XB, Xu XH (2015) Expression analysis of genes encoding mitogen-activated protein kinases in maize provides a key link between abiotic stress signaling and plant reproduction. Funct Integr Genomics 15(1):107–120. https://doi.org/10.1007/s10142-014-0410-3
Takahashi F, Mizoguchi T, Yoshida R, Ichimura K, Shinozaki K (2011) Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol Cell 41(6):649–660. https://doi.org/10.1016/j.molcel.2011.02.029
Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15(1):141–152. https://doi.org/10.1016/j.molcel.2004.06.023
Tuteja N (2007) Abscisic acid and abiotic stress signaling. Plant Signal Behav 2(3):135–138. https://doi.org/10.4161/psb.2.3.4156
Wang F, Jing W, Zhang W (2014a) The mitogen-activated protein kinase cascade MKK1-MPK4 mediates salt signaling in rice. Plant Sci 227:181–189. https://doi.org/10.1016/j.plantsci.2014.08.007
Wang L, Liu Y, Cai G, Jiang S, Pan J, Li D (2014b) Ectopic expression of ZmSIMK1 leads to improved drought tolerance and activation of systematic acquired resistance in transgenic tobacco. J Biotechnol 172:18–29. https://doi.org/10.1016/j.jbiotec.2013.11.006
Wu L, Zu X, Zhang H, Wu L, Xi Z, Chen Y (2015) Overexpression of ZmMAPK1 enhances drought and heat stress in transgenic Arabidopsis thaliana. Plant Mol Biol 88(4–5):429–443. https://doi.org/10.1007/s11103-015-0333-y
Xing Y, Jia W, Zhang J (2008) AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J 54(3):440–451. https://doi.org/10.1111/j.1365-313X.2008.03433.x
Xiong L, Yang Y (2003) Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell 15(3):745–759. https://doi.org/10.1105/tpc.008714
Xu J, Zhang S (2015) Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci 20(1):56–64. https://doi.org/10.1016/j.tplants.2014.10.001
Xu L, Xiang G, Sun Q, Ni Y, Jin Z, Gao S, Yao Y (2019) Melatonin enhances salt tolerance by promoting MYB108A-mediated ethylene biosynthesis in grapevines. Hortic Res 6:114. https://doi.org/10.1038/s41438-019-0197-4
Zaidi I, Ebel C, Belgaroui N, Ghorbel M, Amara I, Hanin M (2016) The wheat MAP kinase phosphatase 1 alleviates salt stress and increases antioxidant activities in Arabidopsis. J Plant Physiol 193:12–21. https://doi.org/10.1016/j.jplph.2016.01.011
Zhang S, Klessig DF (2001) MAPK cascades in plant defense signaling. Trends Plant Sci 6(11):520–527. https://doi.org/10.1016/s1360-1385(01)02103-3
Zhang T, Chhajed S, Schneider JD, Feng G, Song WY, Chen S (2019) Proteomic characterization of MPK4 signaling network and putative substrates. Plant Mol Biol 101(3):325–339. https://doi.org/10.1007/s11103-019-00908-9
Zhou WJ, Leul M (1998) Uniconazole-induced alleviation of freezing injury in relation to changes in hormonal balance, enzyme activities and lipid peroxidation in winter rape. Plant Growth Regul 26(1):41–47. https://doi.org/10.1023/a:1006004921265
Zhou S, Chen Q, Sun Y, Li Y (2017) Histone H2B monoubiquitination regulates salt stress-induced microtubule depolymerization in Arabidopsis. Plant Cell Environ 40(8):1512–1530. https://doi.org/10.1111/pce.12950
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31972358), the Natural Foundation of Shandong Province (Grant No. ZR2018MC022) and Shandong Provincial Key Research and Development Project (Grant No. 2019JZZY010727).
Author information
Authors and Affiliations
Contributions
QS and BL conceived and designed the experiments. XJ, CS, and YY conducted the experiments. XJ and XL analyzed the data. XJ and QS wrote the manuscript. All authors read and approve the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
No conflict of interest exits in the submission of this manuscript, and manuscript is approved by all authors for publication.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Communicated by Henryk Flachowsky.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11240_2021_2218_MOESM1_ESM.tif
Fig. S1 Identification of the transgenic plants of VvMAPK9. (A) Screening of overexpressing strains by antibiotic resistance. (B) Identification of transgenic seedlings by PCR. (C) The expression of VvMAPK9 in the leaves of WT and transgenic plants. Supplementary file1 (TIF 2444 KB)
11240_2021_2218_MOESM2_ESM.tif
Fig. S2 Screening and identification of VvMAPK9-overexpressing grape callus. (A) Wild-type grape callus. (B) Agrobacterium tumefaciens and grape callus were cultured together. (C) Screening transgenic grape callus by antibiotic resistance. (D) Transgenic grape callus was obtained (circled in red). (E) The transgenic grape callus was cultured by screening medium. (F) The expression level of VvMAPK9 in WT and transgenic grape callus. Supplementary file2 (TIF 2894 KB)
Rights and permissions
About this article
Cite this article
Ji, X., Sui, C., Yu, Y. et al. Grape VvMAPK9 positively regulates salt tolerance in Arabidopsis and grape callus through regulating the antioxidative system. Plant Cell Tiss Organ Cult 148, 609–622 (2022). https://doi.org/10.1007/s11240-021-02218-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-021-02218-9