Abstract
Banana (Musa spp.) is an important tropical crop. Banana industry is under biotic and abiotic stresses such as Fusarium wilt, typhoon, cold stress. Genetic engineering offers a powerful strategy to create germplasm of banana with enhanced resistance. The safety of genetically modified organisms has become a bottleneck restricting the popularization and application of genetically modified technology. In this study, a candidate promoter, LEAFY (LFY) for expression and flower initiation in Arabidopsis, was cloned and constructed to ‘Gene-deletor’ vector. Histochemical β-glucuronidase (GUS) staining results showed that the ‘Gene-deletor’ vector driven by LFY promoter could lead to 88.5% excision efficiency from Arabidopsis seeds based on more than 200 T3 progeny examined per event. GUS staining was found to be partially negative in transgenic bananas, however, polymerase chain reaction could still detect the presence of large fragments of the vector. These results suggest that although LFY promoter could not completely drive the ‘Gene-deletor’ vector to achieve the effect of complete elimination of exogenous gene in bananas, its efficiency of eliminating exogenous gene laid a theoretical foundation for cloning banana fruit-specific promoters, that is, ‘non-transgenic’ GM bananas.
Key message
These results suggest that LFY promoter could not completely drive the ‘Gene-deletor’ vector to achieve the effect of complete elimination of exogenous gene in bananas. Nevertheless, a certain effect of exogenous gene elimination laid a theoretical foundation for the next step of screening banana fruit-specific promoters, removing all exogenous genes from banana fruits, and solving the food safety problem of genetically modified bananas.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana (Musa spp.) is not only one of the world’s major fruit crop, but also the staple food of nearly 400 million people in more than 130 countries in Africa, America and Asia, with important economic value (Rustagi et al. 2015; Wei et al. 2016). However, the performance of the banana industry is affected by biotic and abiotic stresses such as Fusarium wilt (Czislowski et al. 2018; Dale et al. 2017), cold stress (Dou et al. 2016), typhoon (Yu et al. 2014). Most banana cultivars are triploid, with long generation times, and sterility, hence, it is difficult to cultivate new resistant banana germplasm by means of cross breeding (Ghag et al. 2014; Tripathi et al. 2008; Yip et al. 2011). In addition, the selection and breeding of new varieties by screening mutants has certain randomness and requires longer periods of time (Hasan et al. 2013). The development of transgenic technology provides new strategies for breeding resistant bananas (Maziah et al. 2007; Sagi et al. 1995; Tripathi et al. 2008). However, the issue of safety with respect to transgenic plants has become a serious constraint for the promotion and application of transgenic technology, and this constraint needs to be resolved at the earliest (Gao et al. 2007; Luo et al. 2007; Tuteja et al. 2012; Wu et al. 2008).
In recent years, a number of biotechnological approaches have emerged to address biosafety issues associated with genetically modified organisms (GMO). These include, use of non-selective marker gene (Khan et al. 2010; Miller et al. 2002), safety marker gene and elimination marker gene (Hohn et al. 2001), male sterility (Muhammad 2005), seed sterility or seedless (Daniell 2002), pollen sterility (Paoletti and Pimentel 1996), chloroplast transformation (Bock 2001; Grevich and Daniell 2007), terminator technology (Daniell et al. 2005). However, these techniques are as yet imperfect or not acceptable (e.g. terminator gene). Due to the low efficiency of exogenous gene elimination, the environmental safety problem of transgenic plants cannot be fundamentally resolved. ‘Gene-deletor’ techniques can facilitate removal of proves that the exogenous genes in transgenic plants from specific organs after the completion of their functions, providing an effective way to solve the biosafety issues of transgenic plants (Li et al. 1992; Luo et al. 2007). This technology utilizes a combination of two site-specific recombinant enzyme components, i.e. the Cre/LoxP system derived from bacterial phages (Chong-Perez et al. 2013; Odell et al. 1990; Zhang et al. 2003) and FLP/FRT from yeast (Woo et al. 2009). The two sets of site-specific synthetic recombinase components are comprehensively utilized to drive the recombinase FLP or Cre to express at a suitable time and site, using a specific promoter (Luo et al. 2007). The recombinase recognizes the fusion recognition site LF, and eliminates the sequence between the two fusion recognition sites (including recombinant enzyme gene sequence) from a specific plant organ cell genome at a specific time (Li et al. 2007). This system is one of the most effective and safe transgenic methods. It is not only more efficient, but also eliminates the exogenous genes in the reproductive materials (Li et al. 2007). Through this system, the exogenous genes of interest in the transgenic plant can be expressed normally in the tissues (such as the root, stem and leaf), thereby achieving their biological functions (such as insect resistance, disease resistance, herbicide resistance). However, the exogenous genes in the organs (e.g., pollen, seeds, fruit) that are not required, can be automatically deleted (Luo et al. 2007).
Therefore, cloning specific promoters of site-specific recombination systems based on traits of crops, trees, fruit trees, vegetables, flowers and grasses are key factor determining the elimination efficiency of GM genes (Luo et al. 2007; Watrud et al. 2004). At present, this system has not been applied to the production of dessert bananas and the feasibility of its application is unknown. It is of great theoretical and practical significance to establish a technical system for the specific elimination of exogenous genes in banana fruits.
In this study, we explored the use of fusion site-specific recombinase system to eliminate exogenous genes in transgenic banana, in order to produce non-transgenic but genetically modified bananas. We cloned and constructed the LEAFY (LFY) promoter-driven recombinase FLP specific-gene elimination vector. Preliminary validation in Arabidopsis showed that the ‘Gene-deletor’ vector we constructed could effectively remove exogenous genes from Arabidopsis fruit. Then, the genetically transformed banana was used to verify the feasibility of the system, and the results showed that the exogenous genes in transgenic bananas had been removed successfully. These findings indicate that the LFY promoter can drive the recombinase FLP to exert a certain role in exogenous gene elimination in bananas. The elimination efficiency of this system needs to be further analyzed by the specific promoter of cloned bananas.
Materials and methods
Plant materials, strains, and transformation vectors
Cavendish (Musa spp. cv. Brazil; AAA Group) banana embryogenic cell suspension and Arabidopsis thaliana Columbia were conserved by the banana group of the Institute of Fruit Tree Research, Guangdong Academy of Agricultural Sciences. Various restriction enzymes, Taq enzymes, In-Fusion ligases and kits were purchased from TaKaRa company, Japan. Kanamycin (Kan), Cefotaxime (Cef), 6-benzyl adenine (6-BA), Indole-3-acetic acid (IAA), 2, 4-Dichlorophenoxyacetic acid (2, 4-D) and α-Naphthyl acetic acid (NAA), biotin and myo-inositol were purchased from Sigma USA. All other chemical reagents were domestically acquired analytical reagents. Bacterial strains of Escherichia coli DH5α and Agrobacterium tumefaciens strain EHA105 were kept in the laboratory as a reserve. Intermediate vector PMD18-T was purchased from TaKaRa company. The pCAMBIA-PAB-polseed-FLP vector was provided by Professor Yi Li of the University of Connecticut. Various media and their formulations used for genetic transformation of bananas are listed in Table S1 (Dou et al. 2016).
LEAFY (LFY) promoter cloning
LFY promoter sequences were downloaded according to the information provided in the A. thaliana genome database (http://www.arabidopsis.org/index.jsp). In order to facilitate the construction of the vector, according to the basic information of pCAMBIA-PAB-polseed-FLP vector, restriction sites of BamHI and SalI, and the protection base were introduced in the upstream and downstream of the primer, respectively, and the extended fragment was about 2354 bp, and the primer sequence was:GSP1-F:5′-GCCGGATCCAGATCTGTTAACGCGAAGAAAGCAAGAAGAAAGTTG-3′ (BamHI), GSP1-R:5′-GCGTCGACAATCTATTTT TCTCTCTCTCTC-3′ (SalI). Genomic DNA of A. thaliana genomic DNA was extracted using TaKaRa column recovery kit instructions (Code No. 9765, TaKaRa, Dalian, China). The PCR amplification was conducted using Arabidopsis genomic DNA as PCR template and the GSP1 stated above as the primer. The amplification procedure was as follows: pre-denaturation at 94 °C for 3 min, 94 °C for 45 s, 60 °C for 30 s, and 72 °C extension for 2 min. In order to reduce the mismatch caused by the amplification process, a total of 30 cycles were performed in this experiment, and the last 72 °C extension was of 10 min duration. According to the instructions of PMD18-T ligase kit (Code No. 6011, TaKaRa, Dalian, China), the amplified target fragment was purified and recovered, and then cloned to the intermediate vector PMD18-T. The positive clones were sent to Sangon Biotech (Shanghai) Co., Ltd. for sequencing, and the recombinant plasmid with correct sequencing was named PT-LFY.
Construction of ‘Gene-deletor’ vector
The PT-LFY vector was double digested with BamHI and SalI, and 2354 bp LFY promoter fragment was recovered. The pCAMBIA-PAB-polseed-FLP vector was also double digested with BamHI and SalI to recover large fragments. The flower primordium determination gene promoter LFY was cloned into the pCAMBIA-PAB-polseed-FLP vector according to the operating instructions of In-Fusion ligase (Code No. 072012, TaKaRa, Dalian, China), and the ligated vector was transformed into E. coli and sent to Sangon Biotech (Shanghai) Co., Ltd. for sequencing. The newly constructed expression vector with correct sequencing was named pCAMBIA-LFY-polseed-FLP. The E. coli plasmid was extracted according to the instruction manual of the plasmid extraction kit (Code No. 9760, TaKaRa, Dalian, China). The newly constructed pCAMBIA-LFY-polseed-FLP plant expression vector was transferred into A. tumefaciens strain EHA105 by heat shock (Huang et al. 2013), for subsequent genetic transformation of Arabidopsis and banana.
Transformation and characterization of transgenic Arabidopsis plants
Arabidopsis seeds were first sterilized with 70% ethanol for more than 10 s then sterilized with 10% sodium hypochlorite for 10 min, washed with sterile water 5 times and seeded on an MS solid medium (Clough and Bent 1998). The seeded medium was incubated at 4 °C and vernalized for 3 days and then shifted at 22 °C for 16 h light/8 h dark conditions. When A. thaliana had growth of 2 or 3 true leaves, they were transplanted to a seedling bowl with vermiculite as the substrate (vermiculite: vegetative soil 1:1) and continued to be cultured. An arched membrane was placed on the surface before culturing in an artificial climate incubator. During the vegetative growth phases, the daily light duration was 16 h (at 20001X) and the temperature was 20 °C. During the reproductive growth phase, the daily light duration was 16 h and the temperature was 20 °C. The light intensity was 20001X. The relative humidity of the incubator was between 70 and 90%. The seedlings were transplanted when they had growth of 4 to 6 true leaves, and were moisturized for 4 to 7 days after transplanting. After the seedlings grew new leaves, appropriate amount of nutrient solution was added every 4 days (2 mol/L KNO3, 2 mol/L CaNO3, 2 mol/L MgSO4·7H20, 1 mol/L NH4NO3, l mol/L KH2PO4, 0.01 mol/L NaMoO4, Iron 1.5 ml/L, 1 mL/L minor); The steps of transformation and characterization of homozygotes using the protocol as previously described (Clough and Bent 1998). In order to verify the elimination effect of ‘Gene-deletor’ on transgenic Arabidopsis thaliana seeds, GUS staining was performed on the root, stem, leaf, anther, fruit and other tissues of transgenic A. thaliana homozygote, and the incidence rates of positive events and non-positive events were calculated. Histochemical β-glucuronidase (GUS) assays were performed as described previously (Jefferson 1987).
Transformation and characterization of transgenic banana plants
The ‘Gene-deletor’ vector was used for transformation of Cavendish banana (Musa spp. cv. Brazil; AAA Group; Accession No. BX_01) embryogenic cell suspensions (ECSs) using the protocol as previously described (Hu et al. 2013). 1 ml PCV ECSs of banana were infected with 10 ml bacterial suspension (OD, 0.2 to 0.5) for 24 h at 26 ± 1 °C with shaking at 50 rpm under dark conditions. The ECSs were then allowed to settle at the bottom and the excess bacterial suspension was removed using a sterile micropipette. The ECSs were washed twice with sterile water, and cultured for 1 month in multiplication medium supplemented with 100 mg/L Kanamycin and 200 mg/L Cefotaxime at 26 ± 1 °C with shaking at 110 rpm in dark conditions.
Genomic DNA was isolated from putative transformed plantlets using a plant DNA extraction kit, according to the manufacturer’s manual (Code No. 9765, TaKaRa, Dalian, China). PCR with NptII gene specific primers GSP2 (GSP2-F:5′-GSP2-F:5′-CTGTGATCTGCCTCAGACTCA-3′,GSP2-R:5′-TCAATCCTTCCTTCTTAATCCTTT-3′) was used to confirm presence or absence of transgene into the plant genome. A 25 μl PCR reaction mixture contained 1 × reaction buffer, 0.2 mM dNTPs, 1.5 mM MgSO4, 1 mM primers, 1 unit of Taq DNA Polymerase and 1 mg of template DNA. The PCR cycles were programmed as follows: 1 cycle at 94 °C for 5 min, followed by 35 cycles at 94 °C for 30 s, 60 °C for 30 s, 72 °C for 60 s, and 1 cycle at 72 °C for 7 min. Southern hybridization assays were performed as described previously (Tripathi et al. 2008). Histochemical β-glucuronidase (GUS) assays were performed as described previously (Jefferson 1987). Positive banana plants were multiplied vegetatively using meristems of in vitro plantlet based on the procedures described in our earlier studies (Dou et al. 2016; Hu et al. 2013). Untransformed Cavendish banana ECSs regenerated offspring were generated as wild control. The rooted plantlets were hardened in the greenhouse and used for further assays.
Analysis of the elimination effect of ‘Gene-deletor’ in transgenic bananas
Five healthy transgenic and untransformed wild control plants were selected and planted in isolated greenhouses. Under normal fertilization and water management, all transgenic plants were able to grow normally, similar to the WT plants. After 10 months of planting, the transgenic and the control plants began to bud, and 7 combs of banana fruit were selected and bagged. The bananas were harvested when they reached 80% maturity, and GUS staining was used to detect the elimination effect of exogenous genes. In order to detect the removal of exogenous genes on a molecular level, TaKaRa column recovery kit was used to extract the DNA from the 5 transgenic lines (Code No. 9765, TaKaRa, Dalian, China). PCR was performed using specific primers according to the method reported by Luo et al. 2007, using the plasmids as positive control; the sequence of the plasmid is GSP3-F:5′- GAACGTGGCGAGAAAGGAAGG-3′,GSP3-R:5′-ACTGACAGAACCGCAACGTTG-3′. The amplification procedure was as follows: pre-denaturation at 94 °C for 3 min, 94 °C for 45 s, 60 °C for 30 s, and 72 °C for 2 min, with a total of 30 cycles, and final extension at 72 °C for 10 min.
Statistical analysis
Experiments were performed according to complete randomized design (CRD). The data reported in the figures are means of the values with standard error (SE) and were examined statistically by ANOVA. Least significant difference (LSD) at the 5% level was analysed by DPS software (version 3.01; Zhejiang University, Hangzhou, China).
Results
Isolation of Arabidopsis LEAFY(LFY)promoter
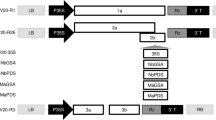
In order to clone the promoter of LEAFY (LFY), the A. thaliana genome DNA was used as a template, and 30 cycles were performed in order to lower the probability of mismatch. After the PCR reaction, all PCR products were subject to agarose gel electrophoresis, and a 5000 bp DNAMarker was used as the molecular standard. As a result, clear bands appeared in about 2.3 kb (Fig. 1a). Then the 2.3 kb DNA amplification band was recovered and purified, and then ligated with the cloning vector PMD18-T. The positive recombinant plasmid with correct sequencing was named PT-LFY. The PCAMBIA-PAB-polseed-FLP vector was digested by the BamHI and SalI, and a clear 1.9 kb fragment was cut (Fig. 1c), which is consistent with the size of 1845 bp PAB5 promoter, indicating that the PAB5 promoter on the pCAMBIA-PAB-polseed-FLP vector was fully excised, and the large fragments vector framework was fully recovered. The PT-LFY was digested through the BamHI and SalI, and the LFY fragments were recovered. The digested vector fragment was ligated with the A. thaliana LFY promoter to form a new recombinant vector. The above-mentioned recombinant ‘Gene-deletor’ vector is digested in the BamHI and SalI, and a clear band produced of the same size as the PCR product (Fig. 1c), indicating that the vector was successfully constructed. The recombinant vector is transformed into Agrobacterium tumefaciens, and the 2.3 kb fragment amplified by PCR (Fig. 1d). The recombinant vector with correct sequencing is named pCAMBIA-LFY-polseed-FLP (Fig. 1e).
Arabidopsis LFY promoter cloning and construction of the ‘Gene-deletor’ vector. a Bands of specific primers of LEAFY promoter, after Arabidopsis thaliana genome PCR amplification. b Bands of original vector pCAMBIA-PAB-polseed-FLP after BamHI and SalI double enzyme digestion and excised PAB promoter. c The large fragments recovered after original vector pCAMBIA-PAB-polseed-FLP double enzyme digestion with BamHI and SalI, and the cloned LFY promoter ligated to the large fragment after enzyme digestion, and then undergoing BamHI and SalI double enzyme digestion to verify whether the LFY promoter ligated with the vector successfully. d PCR positive identification after recombinant vector transformation of Agrobacterium tumefaciens. e Schematic diagram of the newly constructed ‘Gene-deletor’ vector pCAMBIA-LFY-polseed-FLP
Verification of the elimination effect of ‘Gene-deletor’ vector in Arabidopsis thaliana
In order to verify the elimination effect of ‘Gene-deletor’ vector exogenous gene driven by the newly constructed LFY promoter, we first conducted genetic transformation verification in A. thaliana. GUS staining was performed on the roots, stems, leaves, fruit clips and fruit tissues of transgenic Arabidopsis thaliana, and GUS activity was observed in the roots, stems, leaves and fruit clips (Fig. 2a–d), indicating that the newly constructed pCAMBIA-LFY-polseed-FLP vector could not achieve the effect of exogenous gene elimination in the roots, stems, leaves and anthers of A. thaliana. Keeping in mind that the LFY promoter is a key promoter affecting flowering and development of Arabidopsis, we hypothesize that the newly constructed ‘Gene-deletor’ vector might perform a certain eliminating function in late flowering bud differentiation phase (fruits) of plants. To test the above hypothesis, we further stained the transgenic Arabidopsis fruits with GUS, and found that of the 200 Arabidopsis fruits, only 23 showed positive GUS staining and 177 showed negative GUS staining (Fig. 2e). The results indicate that the exogenous genes in the non-stained Arabidopsis thaliana fruits were successfully eliminated, with an elimination effect of 88.5% (Fig. 2f). The above results confirm that our newly constructed ‘Gene-deletor’ vector driven by LFY promoter can achieve certain exogenous gene elimination effect in Arabidopsis thaliana.
Analysis of eliminating effect of ‘Gene-deletor’ vector pBIN19-LFY-FLP in Arabidopsis thaliana. a Transgenic Arabidopsis thaliana homozygous T3 root GUS staining. b GUS staining in the stem of transgenic Arabidopsis thaliana homozygous T3. c GUS staining in leaves of transgenic Arabidopsis thaliana homozygous T3. d GUS staining in fruit pods of transgenic Arabidopsis thaliana homozygous T3. e GUS staining of the fruits of transgenic Arabidopsis thaliana homozygous T3. f The calculated elimination efficiency of exogenous genes in the fruits of transgenic Arabidopsis thaliana homozygous T3. The above experiments were repeated several times. Representative experiments were photographed, and the eliminating efficiency of exogenous genes was calculated
Generation of transgenic banana plants and their molecular analysis
In order to further verify the feasibility of using ‘Gene-deletor’ vector in bananas, ECSs of banana were genetically transformed using A. tumefaciens harboring ‘Gene-deletor’ vector (pCAMBIA-LFY-polseed-FLP) (Fig. 1e). Three subcultures in fresh medium were performed once in 10 days (Fig. 3a). Three months from transformation and selection, whitish embryos emerged on embryo development medium supplemented with 100 mg/L Kanamycin (Fig. 3b). These embryos were later transferred onto germination medium to aid the emergence of first plantlets (Fig. 3c), after which the tiny buds were transferred to multiplication medium. Clusters of small plantlets and white buds were observed after 1 month of culture (Fig. 3d). Single plantlets were isolated and rooted on rooting medium supplemented with 0.1 mg/L NAA (Fig. 3e). Finally, the rooted plantlets were hardened in the greenhouse and used for further analysis (Fig. 3f).
Generation of transgenic banana cv. Cavendish plants using Agrobacterium tumefaciens-mediated transformation. a Embryogenic cell suspension (ECS) and Agrobacterium with ‘Gene-deletor’ vector in it was cultivated together in the proliferating growth medium. b Somatic embryos derived from pCAMBIA-LFY-polseed-FLP transformed cells on embryo development medium at 2 months post selection. c Plantleting after 3 months on embryo germination medium. d Transgenic multiple plantlets on multiplication medium. e Transgenic rooted plantlets on rooting medium. f After rooting of single plantlets on rooting medium, rooted transgenic plants were hardened in the greenhouse
To identify transformed plants, we performed histochemical GUS staining at each cultivation stage. Genomic DNA PCR analysis of the twenty-one GUS positive lines showed that eleven out of them had a single 400-bp fragment derived from the NptII gene coding sequence present in the T-DNA of the binary vector, whereas it was absent in genomic DNA derived from untransformed WT plants (Fig. 4a). The Southern blotting results further confirmed the stable integration of transgenes into Cavendish banana genomes. Five transgenic lines (#1, #5, #11, #13 and #17) with southern blotting and GUS positive lines were used for further analysis, along with the corresponding untransformed WT (Fig. 4b, c). In addition, under a healthy growth condition, we have not observed any phenotypic difference between transgenic lines and the wild-type Cavendish banana (data not shown).
Generation and molecular identification of transgenic banana plants. a PCR confirmation of the Kanamycin-resistant plants using NPTII-specific primers. M molecular marker, WT wild type, - water, the numbers indicate different transgenic lines; P plasmid DNA (used as a positive control). b Southern hybridization to verify the copy number in transgenic banana plants. M, molecular marker; WT wild type, the numbers indicate different transgenic lines; P positive control. c GUS expression in five transgenic lines as assessed by histological GUS assay
Verification of the elimination effect of ‘Gene-deletor’ vector in bananas
The above five well-grown and strong transgenic plants and the wild control plants were planted and isolated in greenhouses, and the bananas were harvested after 1 year or 1 growing season. The fruits harvested from the transgenic and wild control plants were sliced longitudinally and analyzed by GUS staining. Staining results showed that all longitudinal sections of line 1, line 5, line 11, line 13 and line 17 plants were dyed blue to a variable extent, while GUS staining of wild control plants was negative (Fig. 5a). Interestingly, the fruits of the line5 plant were the least stained; fruits on line 17 were stained positive with GUS in half of the longitudinal sections and negative with GUS in the other half (Fig. 5a). PCR results showed that all the five lines obtained the T-DNA fragments of the vector, indicating the existence of the exogenous gene in 5 lines of fruits. However, among them, in addition to 10 kb large fragments obtained from line 1, line 11 and line 13, bands about the size of 100 bp were also obtained, and the 100 bp small fragments were cut out for sequencing. The sequencing results were consistent with the right and left boundaries of the T-DNA vector. The above results fully demonstrate that partial foreign gene elimination occurred in 3 lines, i.e. line 1, line 11 and line 13.
Effect Analysis of ‘Gene-deletor’ vector pBIN19-LFY-FLP in banana fruit. a GUS staining of fruits from 5 transgenic banana lines. b Specific primer molecules tests were used to detect the elimination of exogenous genes in the fruits of five transgenic banana lines. WT wild type, P positive control; the numbers indicate the different transgenic lines. The above experiments were repeated several times. Representative experiments were photographed, and the eliminating efficiency of exogenous genes was calculated
In addition, in order to eliminate the interference of the mosaic on the judgment of eliminating effect, the other half of GUS negative flesh from the line 17 fruits was cut, its genomic DNA was extracted, and specific primers were used for PCR. The results showed that small fragment PCR products were obtained from all the selected materials, but no large fragment PCR products were found. The comparison of banana pulp slice did not show any bands (results not shown), and the results indicated that complete elimination of exogenous genes occurred in part of the line 17 fruits.
Discussion
In recent years, molecular breeding has proved to be one of the most rapid and effective strategies for creating new stress-resistant species (Koch and Kogel 2014; Qi et al. 2018). Generally, exogenous genes are introduced into plants for the purpose of obtaining ideal phenotypes, improving plant stress resistance and improving plant quality (Li et al. 2007). However, these exogenous genes such as antibiotic marker genes, are not necessary for plant growth, and are no longer required for resistance screening after transformation. On the contrary, the presence of these exogenous genes may lead to biosafety and food safety problems (Luo et al. 2007). Therefore, for crops where the fruit is an edible organ (e.g., bananas), it is important to have systems that efficiently and specifically eliminate exogenous genes from the fruit.
In order to eliminate concerns about the biological safety of GM crops and food safety, several molecular strategies have been applied to GM plants in recent years (Daniell 2002; Hills et al. 2007; Kausch et al. 2010), including cotransformation, transposable elements, recombinase deletion and CRISPR/Cas9 techniques (Khan et al. 2010; Li et al. 2007; Luo et al. 2007; Lutz et al. 2011; Naim et al. 2018). However, these existing technologies are mainly used to eliminate resistance genes and reporter genes in transgenic plants, but they cannot effectively and specifically eliminate all exogenous genes, The existence of these exogenous genes may lead to gene escape and ecological hazards (Zuo et al. 2001). Therefore, the technologies stated above cannot be applied in production to solve the safety problems brought by genetically modified crops. In 2007, the Yi Li team at University of Connecticut developed the ‘Gene-deletor’ technology. This technique uses the pollen and seed-specific PAB5 promoters, which automatically eliminate all foreign genes in the pollen and seeds before and after transgenic flowering. This solves the problem of genetic flow of transgenic plants, and solves the concern about the safety of genetically modified food (Li et al. 2007). The efficacy of this technique has been demonstrated in tobacco (Luo et al. 2007).
Recently, Chong-Perez et al. (2013) reported that the feasibility of using developmentally controlled promoters to mediate marker excision in banana. Considering that the LEAFY gene is the key gene affecting plant flowering development, in order to complete the removal of exogenous genes in early fruit formation, in this study the LFY promoter of the flower-determination gene LEAFY from A. thaliana genome was cloned (Fig. 1a), and successfully replaced the PAB5 promoter in the exogenous ‘Gene-deletor’ vector pCAMBIA-PAB-polseed-FLP (Fig. 1b). In order to further verify the eliminating effect of the newly constructed exogenous gene vector pCAMBIA-LFY-polseed-FLP in fruits, we first carried out genetic transformation in A. thaliana. Through GUS staining of the roots, stems, leaves, fruit clip and fruits of transgenic T3 homozygous plants, it was found that only 23 of the 200 Arabidopsis fruits were stained, and 177 Arabidopsis fruits showed no GUS signal, while GUS gene activity was detected in roots, stems, leaves and fruit clips (Fig. 2a–e). These results indicated that the specific promoter LFY in A. thaliana could not remove exogenous genes in roots, stems and leaves after the initiation of vector elimination, but it could effectively remove exogenous genes in Arabidopsis thaliana fruits, with the eliminating effect reaching 88.5% (Fig. 2f).
Since our newly constructed exogenous ‘Gene-deletor’ vector pCAMBIA-LFY-polseed-FLP has good exogenous gene eliminating effect in transgenic Arabidopsis thaliana fruits (Fig. 2), this discovery prompted us to study the feasibility of applying exogenous ‘Gene-deletor’ vector in banana fruits. In order to test our hypothesis, genetic transformation of Cavendish banana was carried out in this paper using the method reported earlier (Dou et al. 2016). The genetically transformed plants were successfully obtained through co-culture, screening, plant regeneration, rooting and other steps (Fig. 3). Five transgenic lines (#1, #5, #11, #13 and #17) were used for further analysis, along with the corresponding untransformed WT. GUS staining was performed on the transgenic banana fruits, and it was found that different levels of exogenous gene eliminating effects were obtained in different parts of the transgenic fruits (Fig. 5a). Luo et al. (2007) reported that after the elimination of exogenous genes, only about 100 bp of vector fragments were left in specific tissues. In this paper, specific primers were used to carry out PCR on the fruits of five transgenic banana lines, and it was found that no small fragments appeared in two lines (line 5 and line 17), indicating that exogenous gene elimination was not completed in the transgenic fruits. Small fragments of about 100 bp appeared in three transgenic lines (line 1, line 11 and line 13), however, the large fragment was still pure (Fig. 5b). These results indicate that the exogenous ‘Gene-deletor’ vector pCAMBIA-LFY-polseed-FLP can achieve certain exogenous gene elimination effect in banana fruits, but cannot completely eliminate exogenous genes. Its elimination effect needs to be further improved.
In conclusion, the application of exogenous ‘Gene-deletor’ technology in bananas has not been previously reported. This paper cloned the LEAFY gene promoter LFY in A. thaliana, and a new exogenous ‘Gene-deletor’ vector pCAMBIA-LFY-polseed-FLP was successfully constructed. Through verification in Arabidopsis, it was found that a better effect of exogenous gene elimination could be achieved in Arabidopsis fruits. Further, it was found that a certain effect of exogenous gene elimination could be achieved in banana fruits, but the exogenous genes could not be completely eliminated. This study laid a theoretical foundation for the next step of screening banana fruit-specific promoters, removing all exogenous genes from banana fruits, and solving the food safety problem of genetically modified bananas.
Abbreviations
- LFY:
-
LEAFY
- GM:
-
Genetically modified
- GMO:
-
Genetically modified organisms
- GUS:
-
Histochemical β-glucuronidase
- PCR:
-
Polymerase chain reaction
- ECS(s):
-
Embryogenic cell suspension(s)
- NptII:
-
Kanamycin gene
- WT:
-
Wild type
- NAA:
-
α-Naphthylacetic
- HPT:
-
Hygromycin B gene
- Cef:
-
Cefotaxime
- BA:
-
6-Benzyl adenine
- IAA:
-
Indole-3-acetic acid
- 2, 4-D:
-
2, 4-Dichlorophenoxyacetic acid
References
Bock R (2001) Transgenic plastids in basic research and plant biotechnology. J Mol Biol 312:425–438
Chong-Perez B, Reyes M, Rojas L, Ocana B, Ramos A, Kosky RG, Angenon G (2013) Excision of a selectable marker gene in transgenic banana using a Cre/lox system controlled by an embryo specifific promoter. Plant Mol Biol 83:143–152
Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Czislowski E, Fraser-Smith S, Zander M, O’Neill WT, Meldrum RA, Tran-Nguyen LTT, Batley J, Aotken EAB (2018) Investigation of the diversity of effector genes in the banana pathogen, Fusarium oxysporum f. sp. cubense, reveals evidence of horizontal gene transfer. Mol Plant Pathol 19:1155–1171
Dale J, James A, Paul J, Khanna H, Smith M, Peraza-Echeverria S, Garcia-Bastidas F, Kema G, Waterhouse P, Mengersen K, Harding R (2017) Transgenic Cavendish bananas with resistance to Fusarium wilt tropical race 4. Nat Commun 8:1–8
Daniell H (2002) Molecular strategies for gene containment in transgenic crops. Nat Biotechnol 20:581–586
Daniell H, Kumar S, Duformantel N (2005) Break through in chlorop last genetic engineering of agronomically important crops. Trends Biotechnol 23:238–245
Dou TX, Hu CH, Sun XX, Shao XH, Wu JH, Ding LJ, Gao J, He WD, Biswas MK, Yang QS, Yi GJ (2016) MpMYBS3 as a crucial transcription factor of cold signaling confers the cold tolerance of banana. Plant Cell, Tissue Organ Cult 125:93–106
Gao XR, Wang GK, Su Q, Wang Y, An LJ (2007) Phytase expression in transgenic soybeans: stable transformation with a vector-less construct. Biotechnol Lett 29:1781–1787
Ghag SB, Shekhawat UK, Ganapathi TR (2014) Host-induced post-transcriptional hairpin RNA-mediated gene silencing of vital fungal genes confers efficient resistance against Fusarium wilt in banana. Plant Biotechnol J 12:541–553
Grevich JJ, Daniell H (2007) Chloroplast genetic engineering: recent advances and perspectives. Crit Rev Plant Sci 24:83–107
Hasan MA, Choudhury RR, Ghosh B, Mandal KK (2013) Screening of banana cultivars to biotic stresses. ISHS Acta Horticulturae 975. In: IV international symposium on tropical and subtropical fruits
Hills MJ, Hall L, Arnison PG, Good AG (2007) Genetic use restriction technologies (GURTs): strategies to impede transgene movement. Trends Plant Sci 12:177–183
Hohn B, Levy AA, Puchta H (2001) Elimination of selection markers from transgenic plants. Curr Opin Biotech 12:139–143
Hu CH, Wei YR, Huang YH, Yi GJ (2013) An efficient protocol for the production of chit42 transgenic Furenzhi banana (Musa spp. AA group) resistant to Fusarium oxysporum. In Vitro Cell Dev Biol 49:584–592
Huang XS, Wang W, Zhang Q, Liu JH (2013) A basic Helix-Loop-Helix transcription factor, PtrbHLH, of Poncirus trifoliata confers cold tolerance and modulates peroxidase-mediated scavenging of hydrogen peroxide. Plant Physiol 162:1178–1194
Jefferson RA (1987) Assaying chimeric plant genes: the GUS fusion system. Plant Mol Biol Rep 5:387–405
Kausch AP, Hague J, Oliver MJ, Hague J, Oliver M, Li Y, Daniell H, Mascia PN, Watrud L, Stewart CN (2010) Transgenic perennial biofuel feed stocks and strategies for bioconfinement. Biofuels 1:163–176
Khan RS, Nakamura I, Mii M (2010) Production and selection of marker-free transgenic plants of Petunia hybrida using site-specific recombination. Biol Plantarum 54:265–271
Koch A, Kogel KH (2014) New wind in the sails: improving the agronomic value of crop plants through RNAi-mediated gene silencing. Plant Biotechnol J 12:821–831
Li Y, Hagen G, Guilfoyle TJ (1992) Altered morphology in transgenic tobacco plants that overproduce cytokinins in specific tissues and organs. Dev Biol 153:386–395
Li Y, Duan H, Smith W (2007) ‘Gene-deletor’: a new tool to address concerns over GE crops. ISB News Report, www.isb.vt.edu/news/2007/news07.jun.htm
Luo KM, Hui D, Zhao DG, Zheng XL, Deng W, Chen YQ Jr, Stewart CN, McAvoy R, Jiang XN, Wu YH, He AG, Pei Y, Li Y (2007) ‘GM-gene-deletor’: fused loxP-FRT recognition sequences dramatically improve the efficiency of FLP or CRE recombinase on transgene excision from pollen and seed of tobacco plants. Plant Biotechnol J 5:263–274
Lutz KA, Azhagiri A, Maliga P (2011) Transplastomics in Arabidopsis: progress toward developing an efficient method. Methods Mol Biol 774:133–147
Maziah M, Sariah M, Sreeraman S (2007) Transgenic banana Rastali (AAB) with β-1,3-glucanase gene for tolerance to fusarium wilt race 1 disease via Agrobacterium-mediated transformation system. Plant Pathol J 6:271–282
Miller M, Tagliani L, Wang N, Berka B, Bidney D, Zhao ZY (2002) High efficiency transgene segregation in co-transformed maize plants using an Agrobacterium tumefaciens 2 T-DNA binary system. Transgenic Res 11:381–396
Muhammad SK (2005) Plant biology: engineered male sterility. Nature 436:783–785
Naim F, Dugdale B, Kleidon J, Brinin A, Shand K, Waterhouse P, Dale J (2018) Gene editing the phytoene desaturase alleles of Cavendish banana using CRISPR/Cas9. Transgenic Res 27:451–460
Odell J, Caimi P, Sauer B, Russell S (1990) Site-directed recombination in the genome of transgenic tobacco. Mol Gen Genet 223:369–378
Paoletti MG, Pimentel D (1996) Genetic engineering in agriculture and the environment. Bioscience 46:665–673
Qi T, Zhu XG, Tan XL, Liu P, Guo J, Kang ZS, Guo J (2018) Host-induced gene silencing of an important pathogenicity factor PsCPK1 in Puccinia striiformis f. sp. tritici enhances resistance of wheat to stripe rust. Plant Biotechnol J 16:797–807
Rustagi A, Jain S, Kumar D, Shekhar S, Jain M, Bhat V, Sarin NB (2015) High efficiency transformation of banana [Musa acuminate L. cv. Matti (AA)] for enhanced tolerance to salt and drought stress through overexpression of a peanut salinity-induced pathogenesis-related class 10 protein. Mol Biotechnol 57:27–35
Sagi L, Panis B, Remy S (1995) Genetic transformation of banana and plantain (Musa spp.) via particle bombardment. Nat Biotechnol 13:481–485
Tripathi L, Tripathi JN, Tushemereirwe WK (2008) Rapid and efficient production of transgenic East African Highland Banana (Musa spp.) using intercalary meristematic tissues. Afr J Biotechnol 7:1438–1445
Tuteja N, Verma S, Sahoo RK, Raveendar S, Reddy IB (2012) Recent advances in development of marker-free transgenic plants: regulation and biosafety concern. J Biosci 37:167–197
Watrud LS, Lee EK, Fairbrother A, Burdick C, Reichman JR, Bollman M, Storm M, King G, Van de Water PK (2004) Evidence for landscape-level, pollen-mediated gene flow from genetically modified creeping bentgrass with CP4 EPSPS as a marker. Proc Natl Acad Sci USA 101:533–538
Wei Y, Hu W, Wang Q, Zeng H, Li X, Yan Y, Reiter RJ, He C, Shi H (2016) Identification, transcriptional and functional analysis of heat-shock protein 90 s in banana (Musa acuminata L.) highlight their novel role in melatonin-mediated plant response to Fusarium wilt. J Pineal Res 62:e12367
Woo HJ, Cho HS, Lim SH, Shin KS, Lee SM, Lee KJ, Kim DH, Cho YG (2009) Auto-excision of selectable marker genes from transgenic tobacco via a stress inducible FLP/FRT site specificrecombination system. Transgenic Res 18:455–465
Wu W, Su Q, Xia XY, Wang Y, Luan LJA (2008) The Suaeda liaotungensis kitag betaine aldehyde dehydrogenase gene improves salt tolerance of transgenic maize mediated with minimum linear length of DNA fragment. Euphytica 159:17–25
Yip MK, Lee SW, Su KC, Lin YH, Chen TY, Feng TY (2011) An easy and efficient protocol in the production of pflp transgenic banana against Fusarium wilt. Plant Biotechnol Rep 5:245–254
Yu B, Chi ZG, Fang Z, Wei SZ, Li BS (2014) Impact of typhoon Wilson on banana production in Guangxi in 2014 and suggestions for post disaster production management. China Trop Agric 5:30–31
Zhang W, Subbarao S, Addae P, Shen A, Armstrong C, Peschke V, Gilbertson L (2003) Cre/lox-mediated marker gene excision in transgenic maize (Zea mays L.) plants. Theor Appl Genet 107:1157–1168
Zuo J, Niu QW, Moller SG, Chua NH (2001) Chemicalregulated, site-specific DNA excision in transgenic plants. Nat Biotechnol 19:157–161
Acknowlegements
This work was supported by the National Natural Science Foundation of China, Nos. 31801843 and 31772267; Natural Science Foundation of Guangdong Province, No. 2017A030310351; Guangdong Academy of Agricultural Sciences Foundation, No. 201815; Project of Science and Technology of Guangzhou, Nos. 201707010154 and 201904020033. Major Special Project of Industry-University-Research Collaborative Innovation in Guangzhou Science and Technology Project No. 201704020031. Science and Technology Plan Project of Guangdong Provice No. 2016B020201007.
Author information
Authors and Affiliations
Contributions
TD, CH and GY conceived of the study and analyzed the data. CH, XS, QY, FB, TD, CL and GD performed the experiment. TD and CH drafted the manuscript. YL and GY revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Additional information
Communicated by Francisco de Assis Alves Mourão Filho.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hu, Ch., Yang, Qs., Shao, Xh. et al. The application of the ‘Gene-deletor’ technology in banana. Plant Cell Tiss Organ Cult 140, 105–114 (2020). https://doi.org/10.1007/s11240-019-01714-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-019-01714-3