Abstract
The explant is an important parameter in plant tissue culture. Most plant tissue culture studies in the literature have focused almost exclusively on the use of explants as a tool for organogenesis or (micro)propagation, and there is limited information that documents the changes that occur in explants in response to cutting during explant preparation just prior to or during culture. To better understand the molecular changes occurring during the wounding process that accompanies the preparation of an explant, this study examined the mRNA transcription profile of 4-week-old potato (Solanum tuberosum L. cv. Desirée) single-node stem segments. To achieve this, RNA-seq analysis was performed to identify significantly differentially expressed genes (DEGs) that existed between uncut in vitro plantlets and cut explants. DEGs corresponding to specific enzyme profiles were broadly connected to cellular processes in the extracellular region, nucleus, and plasma membrane, and were associated with biosynthesis, carbohydrate metabolism and catabolism, cellular protein modification, growth and development, and response to stress. The RNA-seq data of eight highly up- or down-regulated DEGs was validated by RT-qPCR, showing high positive Spearman and Pearson correlation coefficients (0.73 and 0.95, respectively) between SeqMonk LFC and RT-qPCR LFC. These results provide a novel perspective of the changes taking place within explants when they are cut and serve as a valuable basis for the study of explant-related stress in plant tissue culture in other species. In addition, our findings may contribute to an understanding of the success or failure of the tissue culture protocol and subsequent establishment of in vitro cultures by better appreciating the stress-related molecular changes taking place.
Key message
mRNA transcription profiles were used to show how differentially expressed genes in cut stem explants of potato during preparation for plant tissue culture constitutes an abiotic stress, although not fatal.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Without an explant, a plant tissue culture protocol cannot evidently be established. Although this statement may appear obvious, it simply emphasizes the importance of the explant in plant tissue culture. When cultured, an explant is exposed to a host of biotic and abiotic factors that may constitute a stress (Jackson 2003; Şen 2012). The presence of high concentrations of auxin or cytokinin, or even metal cations, in plant tissue culture media may constitute oxidative stress which, alongside the accumulation of lipid peroxidation products, may reduce totipotency and reduce the regenerative ability of cultures, particularly callus, which is a tissue in a dedifferentiated state (Benson and Roubelakis-Angelakis 1994).
The browning or oxidation of tissue cultures, firstly explants and then extending to media, as a result of explant wounding is a well-known phenomenon that involves activation of phenylalanine ammonialyase and peroxiredoxin, the latter being part of the peroxidase antioxidant system (Chen et al. 2012). Wounded plant tissues release ethylene and jasmonates (Lup et al. 2016), as well as auxin, which accumulates at the cut surface and regulates the formation of new vascular tissue (Sauer et al. 2006). In the model plant Arabidopsis thaliana, regeneration of partially cut stems may be epimorphic, i.e., tissue reunion is preceded by extensive cell proliferation (Lup et al. 2016). In cut roots, a suite of transcription factors (TFs) mediate dedifferentiation followed by the establishment of a new root apical meristem, a process that requires auxin (Lup et al. 2016). Auxin is also an essential factor for root organogenesis from other wounded organs, such as stems or leaves, and activates several TFs that then induce cell divisions and restore root identity (Pacurar et al. 2014; Xu 2018). In wounded A. thaliana leaves, genes that are activated at the site of wounding are involved in repair and regeneration (Delessert et al. 2004). Dedifferentiation induced by stress generally allows cells to acquire a stem cell fate and undergo epigenetic reprogramming, which also involves chromatin decondensation and the up-regulation of TFs (Grafi and Barak 2015). This dedifferentiation followed by redifferentiation is a key aspect of de novo shoot organogenesis (Duclercq et al. 2011). Apart from this scanty literature, there are few clues available to plant tissue culture scientists regarding the processes taking place when an explant is cut, and thus wounded.
In a bid to appreciate the changes taking place within an explant in response to cutting, the mRNA expression profile, based on differentially expressed gene (DEG)-based enzyme functions assigned by Blast2GO and KEGG, two powerful tools for transcriptomic analyses (Wang et al. 2009; Lowe et al. 2017), between uncut plantlets and cut explants, in this case, stem single-nodes of 4-wk-old in vitro potato (Solanum tuberosum L.), was assessed.
Materials and methods
Plant material and tissue culture protocol
Following Dobránszki et al. (2017), single-node stem segments with a single leaf cut from 4-week (w)-old in vitro potato plantlets (cv. Desirée) served as the cut explants for this experiment. The control (uncut) treatment were intact 4-wk-old in vitro potato plantlets. All culture conditions, medium and vessels used followed Dobránszki et al. (2017). Except for when the 4-wk-old in vitro plantlets were cut into explants, all other experimental parameters were identical when samples were collected. Samples were collected after opening vessels and gently removing the 4-wk-old in vitro plantlets. Thereafter, whole plantlets (control) as well as explants cut from intact plantlets were immersed immediately into liquid nitrogen. Samples were stored immediately at − 80 °C until RNA extraction and subsequent analysis.
Total RNA isolation and removal of rRNA
Total RNA was purified from six samples (three each from uncut control and cut explant) as three biological replicates using the Direct-zol™ kit (Zymo Research, Irvine, CA, USA) with TRIzol reagent based on the manufacturer’s protocol. After purifying total RNA, three quality control (QC) methods were applied: (1) microcapillary electrophoresis with an Implen n50 nanophotometer (Implen, Munich, Germany) for preliminary quantification; (2) agarose gel electrophoresis to assess RNA degradation and potential contamination; (3) Agilent Bioanalyzer 2100 system (Agilent, Santa Clara, CA, USA). After the three-step QC, mRNA capturing, cDNA library preparation and sequencing were performed by Novogene (Beijing, China), a genome sequencing company. mRNA from the samples was enriched using oligo(dT) beads. For long non-coding libraries, rRNA was removed using the Plant Ribo-Zero rRNA removal kit (Illumina, San Diego, CA, USA) using the manufacturer’s protocol, resulting in mRNA (rRNA-depleted RNA-seq).
mRNA library construction for NGS
Using the Illumina TruSeq Stranded mRNA kit, mRNA was first randomly fragmented by adding fragmentation buffer, then cDNA was synthesized with an mRNA template and a random hexamer primer, after which a custom second-strand synthesis buffer, dNTPs, RNase H and DNA polymerase I were added to initiate second-strand synthesis. The second step involved terminal repair, A ligation and sequencing adaptor ligation, after which the double-stranded cDNA library was completed through size selection and PCR enrichment. The quality of the cDNA library was assessed in three ways: (1) Qubit 2.0 fluorometric quantitation (Thermo Fischer Scientific, Waltham, MA, USA), mainly to assess the library concentration; (2) Agilent Bioanalyzer 2100 (Agilent) to test insert size; (3) AriaMX qPCR (Agilent) to quantify precisely the effective library concentration. The six mRNA libraries derived from the three biological replicates were pooled into two qualified libraries.
Sequencing
The two qualified libraries were fed into the HiSeq 2500 sequencer (Illumina) after pooling according to its effective concentration and expected data volume. For deep sequencing, both libraries were sequenced with 150 bp paired-end reads and the expected data volume was 100 M reads/sample. For each qualified library, three biological replicates were sequenced and pooled into one library.
Bioinformatic processing of RNA-seq datasets
We downloaded the Solanum tuberosum annotated whole genome from the Ensembl Plant database (SolTub 3.0: https://plants.ensembl.org/Solanum_tuberosum/Info/Index) which we used as the reference genome for the RNA-seq datasets. Trimming was performed to remove both poor-quality reads and adapter sequences using TrimGalore v0.5.0 (https://github.com/FelixKrueger/TrimGalore) with default parameters. In addition, 10 bp were removed from the 5′ end of the reads to avoid sequence biases which cause oligo(dT) and random hexamer primers (Hansen et al. 2010). Trimmed reads were aligned to the reference genome with HISAT2 v2.1.0 (Kim et al. 2015) using the paired-end mode. Known splice sites were specified from the splice sites file built from GTF annotation files which were downloaded from the SolTub 3.0 Ensembl Plant database using the HISAT2 python script.
Analysis of RNA-seq datasets
Alignments from HISAT2 were imported into the SeqMonk v1.42.0 (https://github.com/s-andrews/SeqMonk) program, specifying a minimum mapping quality of 60 to select only uniquely aligned reads. The RNA-seq quantitation pipeline was used to quantitate number of reads in the cDNA library at the gene level by counting the number of reads which fall into exons of each gene and correcting for the total number of reads in the samples. The final quantitated values were reads per million reads of input and were log2 transformed. To filter the list of quantitated genes for significant DEGs, the intensity difference filter in SeqMonk, which is based on the χ2 probe, was used to measure differences in gene expression intensity between the two samples (uncut control and cut explant). Throughout this paper, we refer to significantly over- and under-expressed DEGs as up- and down-regulated, respectively.
Functional annotation of significant DEGs
Significantly up- and down-expressed transcribed fragments (transfrags) were selected and blasted against the NCBI database using BlastX-fast in Blast2GO v5.2 (Götz et al. 2008). BlastX-fast was performed against the NCBI nucleotide database (based on the Viridiplanteae) with the minimum E-value score set to 1.0E−06. To assign gene ontology (GO) terms to each annotated sequence, successful blast hits were mapped and annotated using Blast2GO for the significantly up- and down-expressed transfrags with the annotation cut-off threshold set to 55 and the GO level weighting set to 5. After GO mapping, InterProScan was performed and merged to the annotation based on 14 databases (Gene3D, SFLD, SuperFamily, Coils, MobiDBLite, CDD, HAMAP, HMMPanther, HMMPfam, FprintScan, BlastProDom, ProfileScan, HMMTigr, PatternScan) using Blast2GO. KEGG maps (Kanehisa Laboratories; https://www.kegg.jp/kegg/kegg1.html; Kanehisa et al. 2017) were used to search and identify the enzymes and pathways related to DEGs.
Total RNA isolation for RT-qPCR
Total RNA was purified from the two potato samples as three biological replicates each using the Direct-zol™ kit (Zymo Research, Irvine, CA, USA) with TRIzol reagent based on the manufacturer’s protocol. After purifying total RNA, three quality control (QC) methods were applied: (1) microcapillary electrophoresis with an Implen n50 nanophotometer (Implen, Munich, Germany) for preliminary quantification; (2) agarose gel electrophoresis to assess RNA degradation and potential contamination; (3) Agilent Bioanalyzer 2100 system (Agilent Technologies Inc., Santa Clara, CA, USA). For the RT-qPCR analysis, we selected eight DEGs from the RNA-seq datasets (PGSC0003DMG400008781, PGSC0003DMG400010283, PGSC0003DMG400022659, PGSC0003DMG400015183, PGSC0003DMG400008734, PGSC0003DMG400012527, PGSC0003DMG400025668 and PGSC0003DMG400022775) based on the most negative and positive changes in intensity in the SeqMonk logarithmic fold changes (LFC) values. We developed the RT-qPCR primers (Suppl. Table 2) for the chosen DEGs and the two normalising (reference) genes (GAPDH, Actin) with the CLC Main Workbench 7.9.2 (Qiagen, Hilden, Germany).
Validation of DEGs by RT-qPCR
Total RNA (120 ng) was reverse transcribed to cDNA using the FIREScript RT cDNA Synthesis MIX (Solis BioDyne, Tartu, Estonia). RT-qPCR was performed with the 5 × HOT FIREPol EvaGreen qPCR Supermix (Solis BioDyne) on the ABI 7300 real-time PCR system (Thermo Fisher Scientific, USA). For RT-qPCR, four biological replicates from the control (uncut plantlet) and cut explant samples were used. Both RNA-seq and RT-qPCR showed the same direction (up- or down-regulation) of differential expression and differential expression logarithmic fold change, as estimated by RT-qPCR. Spearman and Pearson correlation coefficients were calculated using Microsoft Office 2018 (Microsoft, USA).
Protein analysis by two-dimensional (2-D) gel electrophoresis
Total protein was extracted from two potato samples (control, i.e. uncut plantlet, and cut explant) using the Plant Total Protein Extractor Kit (Sigma-Aldrich, St. Louis, MI, USA). The extracted total plant protein was analyzed by 2-D gel electrophoresis using the Agilent Protein 230 Kit (Agilent, Santa Clara, CA, USA) on an Agilent Bioanalyzer 2100 (Agilent). For the analysis, three biological replicates were used for each sample. We analyzed the total protein samples under reducing (dithiothreitol) and non-reducing (distilled water) conditions.
Results and discussion
Using a powerful transcriptomic tool, RNA-seq, and validated by RT-qPCR (Wang et al. 2009; Lowe et al. 2017), this study aimed to assess the changes in the expression intensity of mRNA in explants in response to cutting in preparation for explant (sub)culture or for any PTC-related procedure. To assist us in answering this question, we employed the model plant potato, and a popular cultivar, Desirée. We found that a number of stress- and development-related DEGs were up-regulated suggesting that the process of cutting, as would be expected, constitutes a stress. However, despite this initial stress, practically speaking, provided that an optimized regeneration protocol is employed, this stress is only temporary, and is fully overcome, allowing normal plantlets to develop (Dobránszki et al. 2017). In other words, in that study, even though the expression of several stress-related antioxidant enzymes (ascorbate peroxidase, glutathione reductase, superoxide dismutase) was detected in wounded explants, ultimately, regeneration was not affected, resulting in shoot production in 100% of stem single-node explants.
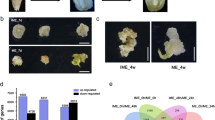
In order to assess the global changes in RNA expression profiles, the expression intensity of 40,430 genes was assessed. From these, after significantly (P > 0.05) up- and down-regulated DEGs (i.e., with significantly higher and lower expression intensity, respectively) were compared using GO annotation (Suppl. Table 1), 189 genes were found to be up-regulated while only six were down-regulated (i.e., with significantly lower expression intensity) in cut explants compared to uncut plantlets (Fig. 1a, b). Significantly down-regulated DEGs are highlighted in green in Suppl. Table 1 while a heat map shows the expression intensity of DEGs between the two samples (Suppl. Fig. 1).
RNA-seq/DEG analysis was used to assess the biological, cellular and molecular processes that were affected in response to cutting. When comparing the significantly up- and down-regulated processes, eight biological processes were down-regulated and five were up-regulated, two were down-regulated and three were up-regulated for cellular processes, and five molecular processes were both down- and up-regulated (Suppl. Fig. 2; Table 1).
What metabolic and cellular functions were up- or down-regulated in response to cutting? When the uncut plantlets and cut explants were compared, emphasis was placed on significantly up- or down-regulated DEGs related to amino acids, carbohydrates, fatty acids, vitamins, nucleotides, as well as growth and development, and stress. Except for amino sugar and nucleotide sugar metabolism, all other processes were up-regulated (Table 2; Fig. 2). Up- and down-regulated DEGs, when annotated to enzymes assigned by Blast2Go and KEGG, resulted in the increased production of products related to defense, stress response, and wound healing, but decreased production of N-acetylglucosamine (Fig. 2). When whole 4-wk-old plants were cut into single-node explants to establish a new subculture, 11 transcription factors were significantly up-regulated and only one was significantly down-regulated (Table 3).
Enzymes coded by significantly up- or down-regulated DEGs related to amino acids, carbohydrates, fatty acids, vitamins, nucleotides, as well as growth and development, and stress. ADP, adenosine diphosphate; β-ALA, β-alanine; AldA, aldarate; AsA, ascorbate; BS, biosynthesis; GA, gibberellic acid; GABA, γ-aminobutyric acid; GAL, galactose; GLY, glycine; JA, jasmonic acid; α-LA, α-linoleic acid; MeJA, methyl jasmonate; MET, metabolism; NAG, N-acetylglucosamine; 10-OPDA, 10-oxo-11,15-phytodienoic acid (“death” acid); PABA, aminobenzoate; PHE, phenyalanine; SER, serine; TA, traumatic acid; THI, thiamine; TRP, tryptophan; TYR, tyrosine. Inner circle (green), process related to growth; middle circle (blue), process related to growth and stress; outer circle (orange), process related to stress. Red text = significantly down-regulated; black text = significantly up-regulated
For RNA-seq/DEG data validation, we used RT-qPCR and 2-D gel electrophoresis. Using RT-qPCR, we detected all of the chosen DEGs as true positive up- or down-regulated DEGs (Suppl. Table 3; Suppl. Fig. 4). The Spearman and Pearson correlation coefficients between SeqMonk LFC and RT-qPCR LFC were 0.73 and 0.95, respectively. These high correlation coefficients indicate a high positive correlation between SeqMonk LFC and RT-qPCR LFC.
Total protein electrophoresis revealed differences between control and treated samples (Suppl. Fig. 5; Suppl. Table 4).
When an explant is cut, like other plant tissues, a host of biochemical and genetic changes occur, the antioxidant system is activated and ethylene and jasmonates are released (Lup et al. 2016). Jasmonates, including jasmonic acid (JA), are derived from 12-oxo-phytodienoic acid (12-OPDA), a cyclopente(a)none, and its derivatives, and are generally involved in protection against biotic stress and are also involved in plant development (Thoma et al. 2003; Christensen et al. 2016). In our study, in response to cutting, DEGs related to JA precursors as well as 10-OPDA, a “death acid” involved in the protection of cells and inhibition of cell death (Christensen et al. 2016), were up-regulated (Fig. 2). 10-OPDA is a precursor to MeJA and to traumatic acid, a potent agent for wound healing (English et al. 1939). Jasmonates are typically released in response to wounding (Lup et al. 2016), and this was additionally evidenced by the up-regulation of DEGs related to methyl jasmonate (MeJA) (Fig. 2). Ethylene is also released in response to wounding (Lup et al. 2016), and this was demonstrated by the activation of seven ethylene-responsive TFs in cut potato stem explants (Table 3).
In wounded A. thaliana leaves of 5-w-old in vitro plantlets, genes coding for signal transduction and regulatory factors, or genes controlled by MeJA, were predominantly expressed (Delessert et al. 2004). Delessert et al. (2004) also detected the activation of genes coding for lignin biosynthesis, as we also observed in cut potato stem explants where DEGs coding for four types of lignin were up-regulated (Fig. 2). Lignin biosynthesis is a typical response to abiotic and biotic stresses (Moura et al. 2010). In addition, Delessert et al. (2004) found that 21% of wound-responsive genes coded for elements related to transcription regulation and signal transduction, including TFs such as WRKY, which are involved in stress and development (Phukan et al. 2016; Zhang et al. 2017), as we also noted in wounded potato stems (Table 3). In their study, Delessert et al. (2004) also noted that 9% of up-regulated genes were related to protein synthesis, 30% related to metabolism and energy supply, including sugar metabolism, and 50% were related to fatty acid or carbon metabolism, and wounded potato stem explants showed strong similarities (Table 2; Fig. 2). Unfortunately, Delessert et al. (2004) did not classify 25% of wound-responsive genes that were related to stress, detoxification or defense while 40% of wound-responsive genes had an unclassified or unknown function. It is not unusual to find an overlap between wound-responsive genes and genes activated by biotic stress (Delessert et al. 2004).
After cutting, DEGs related to carotenoid and phenylpropanoid biosynthesis as well as α-linolenic acid metabolism were up-regulated (Fig. 2; Suppl. Fig. 3). In carotenoid biosynthesis, xanthoxin, an ABA precursor (Xiong et al. 2002), would be produced when 9-cis-epoxycarotenoid dioxygenase is up-regulated. Xanthoxin is related to stress response in plants (Ruiz-Sola and Rodríguez-Concepción 2012). In the liverwort, upregulation of phenylpropanoid biosynthesis was connected to the up-regulation of phenylalanine metabolism in response to cutting-induced stress (Yoshikawa et al. 2018). Gibberellin 2-β-dioxygenase participates at eight points of the diterpenoid biosynthesis pathway and up-regulation of its DEGs could lead to the production of gibberellin catabolites (Fig. 2; Suppl. Fig. 3).
Either in their role as a regulator, or in signaling, amino acids form part of the stress response in plants (Zeier 2013). When explants were cut, DEGs related to alanine, aspartate and glutamate, β-alanine, and histidine metabolism were up-regulated, which could result in increased production of glutamate and aspartate, and the signaling amino acid, γ-aminobutyric acid (GABA). GABA, like JA, plays a signaling role in response to stress and in growth and development (González-Gallegos et al. 2015; Ramesh et al. 2017). Up-regulation of DEGs related to glycine, serine and threonine metabolism affects tryptophan metabolism, just like the up-regulation of DEGs related to phenyalanine, tryptophan and tyrosine biosynthesis at several points within the pathway (Table 2). Moreover, up-regulation of DEGs related to phenyalanine, tryptophan and tyrosine biosynthesis might result in an increase in indole diterpene alkaloid and phenypropanoid biosynthesis via increased production of 4-hydroxy phenylpyruvate (Fig. 2; Suppl. Fig. 3), thereby activating the secondary antioxidant system and increasing the level of NADPH (Yoshikawa et al. 2018). Upregulation of DEGs coding for histidine decarboxylase (Suppl. Fig. 3) in histidine metabolism might increase histamine production after cutting (Fig. 2). Histamine is related to the stress response (Kumar et al. 2016). Up-regulation of a glutamate decarboxylase DEG (Suppl. Fig. 3) might result in increased production of taurine and hypotaurine (Fig. 2), which are membrane-stabilizing antioxidants (Jong et al. 2012), or of GABA while up-regulation of linoleic acid metabolism-related DEGs would result in the production of JA precursors (Fig. 2; Suppl. Fig. 3).
Increased production of different sugars may be connected to increased energy demand and signaling of stress response after cutting (Gupta and Kaur 2005). After explant cutting, a galactose metabolism-related DEG was up-regulated, which would result in galactinol production and finally the production of galactose, glucose and fructose through raffinose production. Galactinol and raffinose are antioxidant and osmoprotective agents that form in response to different environmental stresses (Nishizawa et al. 2008). Down-regulation of DEGs related to amino sugar and nucleotide sugar metabolism might result in a change in the production of N-acetyl glucosamine (Fig. 2), which is involved in stress response, growth and development in plants (Konopka 2012).
Changes to glycerolipid content and composition of cell membranes, which depends on the length and degree of saturation of fatty acids, is a typical reaction to different abiotic stresses (Higashi et al. 2015). Upregulation of an acylglycerol lipase DEG in glycerolipid metabolism may regulate the network of fatty acids (Fig. 2; Suppl. Fig. 3).
DEGs in pathways related to vitamins (aminobenzoate degradation, ascorbate and aldarate, and thiamine metabolism) were up-regulated in response to cutting (Fig. 2; Suppl. Fig. 3). The eventual production of thiamine phosphate and finally thiamine would increase as a result of nucleoside-triphosphate phosphatase up-regulation while up-regulation of nitrophenyl phosphatase might increase the level of 4-nitrophenol leading to the degradation of benzoate. Up-regulation of the DEGs coding for L-ascorbate oxidase may increase the level of L-dehydro-ascorbate, which is a marker of the activation of the ascorbate–glutathione pathway in response to cutting stress (Hanson et al. 2016).
Conclusions
The objective of this study was to assess whether the preparation of an explant for plant tissue culture, or the establishment of an in vitro culture, constituted a stress and how it can change the mRNA expression profile. Current theoretical models and evidence from model plants in the plant-based stress-related literature suggest that cutting is a wound-inducing stress. From those studies, the DEGs and enzyme systems potentially related to those DEGs, as well as the stress-related compounds that were shown in this study to be up-regulated in response to explant cutting, confirm that the process itself is a stress. RT-qPCR of eight highly up- or down-regulate DEGs validated the RNA-seq analyses. Although this finding might appear to be self-evident, it constitutes the first transcriptomic confirmation of this possibility in plant stress. These findings may spur other researchers to examine the stress-related profile of explants in different plant species and with additional evidence from other plants. It would then be possible to appreciate the mechanism by which an explant suffers a stress, then either suffers irreparable shock and dies, or recovers to either redifferentiate (stem cell fate) or dedifferentiate (callus induction).
Changes in the mRNA expression profile in response to cutting and their connection to stress response and growth and developmental processes have been described in the present study. Systematic signaling responses triggered by wounding stress while preparing explants, i.e. by cutting, are another form of abiotic stress, are also common to cold, heat, drought or salt stress, while the wounding response is similar to that caused by pathogen attack (Delessert et al. 2004; Zhu 2016). Information about the changes and intensity of gene expression after explant cutting may contribute to revealing mechanisms that promote the survival and regeneration of explants and plantlets after stress. The described molecular (transcriptomic) responses may be useful when designing molecular breeding and transgenic engineering strategies to improve abiotic stress tolerance and fortify the environmental adaptation of plants (Pereira 2016).
References
Benson EE, Roubelakis-Angelakis KA (1994) Oxidative stress in recalcitrant tissue cultures of grapevine. Free Radic Biol Med 16:355–362. https://doi.org/10.1016/0891-5849(94)90037-X
Chen G, Chen D-Y, Wang T, Xu C-J, Li L (2012) Analysis of the proteins related to browning in leaf culture of Phalaenopsis. Sci Hortic 141:17–22. https://doi.org/10.1016/j.scienta.2012.03.027
Christensen SA, Huffaker A, Hunter CT, Alborn HT, Schmelz EA (2016) A maize death acid, 10-oxo-11-phytoenoic acid, is the predominant cyclopentenone signal present during multiple stress and developmental conditions. Plant Signal Behav 11:e1120395. https://doi.org/10.1080/15592324.2015.1120395
Delessert C, Wilson IW, Van Der Straeten D, Dennis ES, Dolferus R (2004) Spatial and temporal analysis of the local response to wounding in Arabidopsis leaves. Plant Mol Biol 55:165–181. https://doi.org/10.1007/s11103-004-0112-7
Dobránszki J, Asboth G, Homoki D, Bíró-Molnár P, Teixeira da Silva JA, Remenyik J (2017) Ultrasonication of in vitro potato single node explants: activation and recovery of antioxidant defence system and growth responses. Plant Physiol Biochem 121:153–160. https://doi.org/10.1016/j.plaphy.2017.10.022
Duclercq J, Sangwan-Norreel B, Catterou M, Sangwan RS (2011) De novo shoot organogenesis: from art to science. Trends Plant Sci 16:597–606. https://doi.org/10.1016/j.tplants.2011.08.004
English J Jr, Bonner J, Haagen-Smit AJ (1939) Structure and synthesis of a plant wound hormone. Science 90:329. https://doi.org/10.1126/science.90.2336.329
González-Gallegos E, Laredo-Alcalá E, Ascacio-Valdés J, de Rodríguez DJ, Hernández-Castillo FD (2015) Changes in the production of salicylic and jasmonic acid in potato plants (Solanum tuberosum) as response to foliar application of biotic and abiotic inductors. Am J Plant Sci 6:1785–1791. https://doi.org/10.4236/ajps.2015.611179
Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36:3420–3435. https://doi.org/10.1093/nar/gkn176
Grafi G, Barak S (2015) Stress induces cell dedifferentiation in plants. Biochim Biophys Acta 1849:378–384. https://doi.org/10.1016/j.bbagrm.2014.07.015
Gupta AK, Kaur N (2005) Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J Biosci 30:761–776. https://doi.org/10.1007/BF02703574
Hansen KD, Brenner SE, Dudoit S (2010) Biases in Illumina transcriptome sequencing caused by random hexamer priming. Nucleic Acids Res 38:e131–e131. https://doi.org/10.1093/nar/gkq224
Hanson AD, Beaudoin GA, McCarty DR, Gregory JF III (2016) Does abiotic stress cause functional B vitamin deficiency in plants? Plant Physiol 172:2082–2097. https://doi.org/10.1104/pp.16.01371
Higashi Y, Okazaki Y, Myouga F, Shinozaki K, Saito K (2015) Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci Rep 5:10533. https://doi.org/10.1038/srep10533
Jackson MB (2003) Aeration stress in plant tissue cultures. Bulg J Plant Physiol special issue: 96–109
Jong CJ, Azuma J, Schaffer S (2012) Mechanism underlying the antioxidant activity of taurine: prevention of mitochondrial oxidant production. Amino Acids 42:2223–2232. https://doi.org/10.1007/s00726-011-0962-7
Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K (2017) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. https://doi.org/10.1093/nar/gkw1092
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360. https://doi.org/10.1038/nmeth.3317
Konopka JB (2012) N-Acetylglucosamine functions in cell signaling. Scientifica 2012:489208. https://doi.org/10.6064/2012/489208
Kumar R, Jiwani G, Pareek A, Kumar ST, Khurana A, Sharma AK (2016) Evolutionary profiling of group II pyridoxal-phosphate-dependent decarboxylases suggests expansion and functional diversification of histidine decarboxylases in tomato. Plant Genome 9(1):1–15. https://doi.org/10.3835/plantgenome2015.07.0057
Lowe R, Shirley N, Bleackley M, Dolan S, Shafee T (2017) Transcriptomics technologies. PLoS Comput Biol 13:e1005457. https://doi.org/10.1371/journal.pcbi.1005457
Lup SD, Tian X, Xu J, Pérez-Pérez JM (2016) Wound signaling of regenerative cell reprogramming. Plant Sci 250:178–187. https://doi.org/10.1016/j.plantsci.2016.06.012
Moura JCMS, Bonine CAV, de Oliveira Fernandes Viana J, Dornelas MC, Mazzafera P (2010) Abiotic and biotic stresses and changes in the lignin content and composition in plants. J Integr Plant Biol 52:360–376. https://doi.org/10.1111/j.1744-7909.2010.00892.x
Nishizawa A, Yabuta Y, Shigeoka S (2008) Galactinol and raffinose constitute a novel function to protect plants from oxidative damage. Plant Physiol 147:1251–1263. https://doi.org/10.1104/pp.108.122465
Pacurar DI, Perrone I, Bellini C (2014) Auxin is a central player in the hormone cross-talks that control adventitious rooting. Physiol Plant 151:83–96. https://doi.org/10.1111/ppl.12171
Pereira A (2016) Plant abiotic stress challenges from the changing environment. Front Plant Sci 7:1123. https://doi.org/10.3389/fpls.2016.01123
Phukan UJ, Jeena GS, Shukla RK (2016) WRKY transcription factors: molecular regulation and stress responses in plants. Front Plant Sci 7:760. https://doi.org/10.3389/fpls.2016.00760
Ramesh SA, Tyerman SD, Gilliham M, Xu B (2017) γ-Aminobutyric acid (GABA) signalling in plants. Cell Mol Life Sci 74:1–27. https://doi.org/10.1007/s00018-016-2415-7
Ruiz-Sola MÁ, Rodríguez-Concepción M (2012) Carotenoid biosynthesis in arabidopsis: a colorful pathway. In: The Arabidopsis Book, e0158. https://doi.org/10.1199/tab.0158
Sauer M, Balla J, Luschnig C, Wiśniewska J, Reinöhl V, Friml J, Benková E (2006) Canalization of auxin flow by Aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev 20:2902–2911. https://doi.org/10.1101/gad.390806
Şen A (2012) Oxidative stress studies in plant tissue culture. In: El-Missiry MA (ed) Antioxidant enzyme. IntechOpen Limited, London, pp 59–88. https://doi.org/10.5772/48292
Thoma I, Loeffler C, Sinha AK, Gupta M, Krischke M, Steffan B, Roitsch T, Mueller MJ (2003) Cyclopentenone isoprostanes induced by reactive oxygen species trigger defense gene activation and phytoalexin accumulation in plants. Plant J 34:363–375. https://doi.org/10.1046/j.1365-313X.2003.01730.x
Wang Z, Gerstein M, Snyder M (2009) RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. https://doi.org/10.1038/nrg2484
Xiong L, Schumaker KS, Zhu J-K (2002) Cell signaling during cold, drought, and salt stress. Plant Cell Suppl 14:S165–S183. https://doi.org/10.1105/tpc.000596
Xu L (2018) De novo root regeneration from leaf explants: wounding, auxin, and cell fate transition. Curr Opin Plant Biol 41:39–45. https://doi.org/10.1016/j.pbi.2017.08.004
Yoshikawa M, Luo W-F, Tanaka G, Konishi Y, Matsuura H, Takahashi K (2018) Wounding stress induces phenylalanine ammonia lyases, leading to the accumulation of phenylpropanoids in the model liverwort Marchantia polymorpha. Phytochemistry 155:30–36. https://doi.org/10.1016/j.phytochem.2018.07.014
Zeier J (2013) New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ 36:2085–2103. https://doi.org/10.1111/pce.12122
Zhang Z, Mao C, Shi Z, Kou X (2017) The amino acid metabolic and carbohydrate metabolic pathway play important roles during salt-stress response in tomato. Front Plant Sci 8:1231. https://doi.org/10.3389/fpls.2017.01231
Zhu J-K (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324. https://doi.org/10.1016/j.cell.2016.08.029
Acknowledgements
This research was financed by the Higher Education Institutional Excellence Programme of the Ministry of Human Capacities in Hungary, within the framework of the Biotechnology thematic programme of the University of Debrecen (20428-3/2018/FEKUTSTRAT). Open access funding provided by University of Debrecen (DE). The authors also wish to thank Dr. Judit Remenyik (University of Debrecen) for useful discussion. The study and submission for publication was approved by the University of Debrecen (BPTR/DEENK/0004/2018). Finally, the authors wish to thank Kanehisa Laboratories for copyright permission (180345) to use KEGG maps used in Suppl. Fig. 3.
Author information
Authors and Affiliations
Contributions
JD conceived the experiments. JD, NH and JATdS designed the experiments. JD established in vitro cultures. NH and AG conducted bioinformatics analyses. JD, NH, AG and JATdS analysed the data, and co-wrote all versions of the paper. JD, NH, AG and JATdS take responsibility for the content of the paper.
Corresponding authors
Ethics declarations
Conflict of interest
All four authors (JATdS, NH, AG and JD) declare no conflict of interest.
Data availability
The raw Illumina mRNA sequences were submitted to the NCBI and the processed data were deposited under GEO ID GSE123037, BioProject ID PRJNA507365, and SRA ID SRP171017 for the two samples: GSM3494235 and GSM3494236.
Additional information
Communicated by José M. Seguí-Simarro.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Teixeira da Silva, J.A., Hidvégi, N., Gulyás, A. et al. mRNA transcription profile of potato (Solanum tuberosum L.) in response to explant cutting. Plant Cell Tiss Organ Cult 138, 143–152 (2019). https://doi.org/10.1007/s11240-019-01613-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11240-019-01613-7