Abstract
A novel Eimeria Schneider, 1875 species is described from an Australian pied oystercatcher Haematopus longirostris Vieillot, in Western Australia. The pied oystercatcher was admitted to the Kanyana Wildlife Rehabilitation Centre (KWRC), Perth, Western Australia in a poor body condition, abrasion to its right hock and signs of partial delamination to its lower beak. Investigation into potential medical causes resulted in a faecal sample being collected and screened for gastrointestinal parasites. Unsporulated coccidian oocysts were initially observed in the faeces and identified as Eimeria upon sporulation. The sporulated oocysts (n = 20) are ellipsoidal, 20–21 × 12–13 μm in shape and have thick bi-layered walls which are c.2/3 of the total thickness. Micropyle is present, robust and protruding, and occasionally has a rounded polar body attached to the micropyle. Within the oocyst, a residuum, in addition, two to five polar granules are present. There are four ellipsoidal sporocysts 9–11 × 5–6 μm with flattened to half-moon shaped Stieda bodies. Sub-Stieda body and para-Stieda body are absent. The sporocysts contain sporocyst residuums composed of a few spherules scattered among the sporozoites. Within the sporozoites, anterior and posterior refractile bodies are present, but the nucleus is indiscernible. To further characterise the novel Eimeria species from H. longirostris, molecular analysis was conducted at the 18S ribosomal RNA locus, using PCR amplification and cloning. Two cloned sequences from the novel Eimeria were compared with those from other Eimeria spp. with the highest genetic similarity of 97.6% and 97.2% from Clone 1 and 2, respectively with Eimeria reichenowi (AB544308) from a hooded crane (Grus monacha Temminck) in Japan. Both sequences grouped in a clade with the Eimeria spp. isolated from wetland birds, which include Eimeria paludosa (KJ767187) from a dusky moorhen (Gallinula tenebrosa Gould) in Western Australia, Eimeria reichenowi (AB544308) and Eimeria gruis (AB544336) both from hooded cranes. Based on the morphological and molecular data, this Eimeria sp. is a new species of coccidian parasite and is named Eimeria haematopusi n. sp. after its host H. longirostris.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The Australian pied oystercatcher (Haematopus longirostris) is a medium-sized black and white wading bird with distinctive features including a bright orange-red bill, red legs, red eye rings and eyes (Pizzey and Knight, 2007). It is found along much of the Australian and Tasmanian coastline, the south coast of New Guinea and the coasts of the Aru and Kai Islands in Indonesia (Hockey et al., 2020).
The Australian pied oystercatcher belongs to the Order Charadriiformes and the Suborder Charadrii (waders) and is a member of the Haematopodidae family. There are 12 species of oystercatcher in the Haematopus genus (Winkler et al., 2020) that include the pied oystercatcher (H. longirostris Vieillot), Eurasian oystercatcher (H. ostralegus Linnaeus), South Island oystercatcher (H. finschi Martens), Chatham oystercatcher (H. chathamensis Hartert), variable oystercatcher (H. unicolor Forster), sooty oystercatcher (H. fuliginosus Gould), American oystercatcher (H. palliatus Temminck), African oystercatcher (H. moquini Bonaparte), Canarian oystercatcher (H. meadewaldoi Bannerman), blackish oystercatcher (H. ater Vieillot), magellanic oystercatcher (H. leucopodus Garnot) and the black oystercatcher (H. bachmani Audubon) (Winkler et al., 2020).
Taxonomical classification places the oystercatchers genetically closest to the ibisbill (Ibidorhynchidae). Together, they form a clade that is most closely related to the avocets and stilts (Recurvirostridae) (Winkler et al., 2020).
Eimeria spp. are the eimeriid coccidia with the greatest parasitic diversity, being found in all groups of vertebrates, in addition to invertebrates (Duszynski, 2021). Isospora spp. are also widely diverse, but are predominantly found in Passeriformes, but also in other orders such as Strigiformes and Struthioniformes (Berto et al., 2011; 2023; Medina et al., 2019; Woodyard et al., 2019; Coker et al., 2023). Within the Charadriiformes, fewer than 22 species of Eimeria (the largest of the Eimeriidae) have been documented to infect members of this order. The Eimeria species previously documented include Eimeria burchinici (Dzerzhinskii and Kairooaev, 1989) in a stone curlew (Burhinus oedicencus Linnaeus), six Eimeria species from plovers (Family Charadriidae), five Eimeria species from the sandpipers (Family Scolopacidae), Eimeria stercorariae Galli-Valerio, 1940 from a parasitic jaeger (Stercorarius parasiticus (Linnaeus)), eight Eimeria species from gulls and terns (Laridae family) and Eimeria fraterculae Leighton and Gajadhar, 1986 from a common puffin (Fratercula arctica (Linnaeus)) (Duszynski et al., 2001).
To date, there is only one species of Eimeria described from unsporulated oocysts from the Haematopus genus, namely Eimeria haematopi, isolated from the kidneys of a Eurasian oystercatcher (H. ostralegus) (Gottschalk and Prange, 2011). In a later study by Siebert et al. (2012) the detection of coccidian infection in histological kidney sections of two Eurasian oystercatchers (H. ostralegus) was evident during the investigation of the health status of seabirds along the North Sea coast of Germany. Unfortunately, no oocysts were isolated or characterized from the study.
In this study, we characterize a new species of Eimeria from a wild Australian pied oystercatcher (H. longirostris), both morphologically and genetically, and propose the species name Eimeria haematopusi n. sp.
Materials and methods
Sample collection
An Australian pied oystercatcher (H. longirostris) was admitted to the Kanyana Wildlife Rehabilitation Centre (KWRC), Perth, Western Australia, in August 2019 after it had been found on the ground in an unusual location after a crash landing. Initial examination showed that the bird was alert and responsive, but it presented in an extremely poor body condition (1/5). It had reduced grip in both feet and an abrasion to its right hock. The lower beak showed signs of partial delamination. To investigate potential medical causes a faecal sample was collected into a sterile, pre-labelled collection vial so that it could be screened for gastrointestinal parasites. The faecal sample was stored at 4°C until microscopy was performed. Despite intensive treatment, the bird died a few days later.
Ethical review and approval were waived for this study based on the Australian code for the care and use of animals for scientific purposes (2013, 8th Edition and updated in 2021) and Animal Welfare Act 2002 (Western Australia). The faecal samples were collected from the debilitated oystercatcher by volunteers at the KWRC for routine diagnostic purposes with the permission of the Board of the KWRC.
Morphological observation
Direct microscopic examination was performed on the faecal sample and a few unsporulated coccidian oocysts were seen. A portion of the faeces was emulsified in a 2% (w/v) potassium dichromate solution (K2Cr2 O7) and stored at 4°C until transport to Murdoch University. Once at Murdoch University, a faecal float was performed on the sample mixture using a saturated sodium chloride and 50% sucrose solution (w/v). A shallow layer (less than 1cm) of the sample mixture was also poured into a Petri dish, placed inside a cupboard, and allowed to sporulate at room temperature (20–22°C). An Olympus DP71 digital micro-imaging camera was used to observe the sporulated oocysts, using the 100× oil immersion objective. Images were captured using a Nomarski contrast imaging system.
DNA extraction
Total DNA was extracted from 200 mg of each faecal sample using a Power Soil DNA Kit (MolBio, Carlsbad, California) with some modifications as described by Yang et al. (2015). Briefly, samples were subjected to four cycles of freezing/thawing in liquid nitrogen and boiling water to ensure efficient lysis of oocysts, before being processed using the manufacturer’s protocol.
Polymerase Chain Reaction (PCR) amplification and cloning of 18S rRNA
Amplification of the Eimeria 18S rRNA region was performed using PCR according to the protocols described by Yang et al. (2016). To identify singular species, cloning of the PCR product was performed as described previously by Yang et al. (2013a) while sequencing and DNA purification were conducted according to previously described protocols (Yang et al., 2013b).
Phylogenetic analysis
The two18S rRNA sequences from E. haematopusi n. sp. were aligned with other closely related Eimeria and Isospora sequences retrieved from GenBank (Benson et al., 2005) via BLAST (Altschul et al., 1990) searches. Phylogenetic trees of 18S rRNA sequences were constructed using MEGA-X (Kumar et al., 2018) with the most appropriate nucleotide substitution models (TN93 + G + I).
The robustness of nodes within the resulting trees was inferred from 1000 cycles of bootstrap resampling.
Line drawing
Line drawings were edited using two software applications from CorelDRAW® (Corel Draw Graphics Suite, Version 2020, Corel Corporation, Canada), i.e., Corel DRAW and Corel PHOTO-PAINT (Yang et al., 2021).
Results
Low numbers of unsporulated coccidian oocysts were detected by light microscopy and faecal flotation. Upon sporulation, the oocysts were characterised as an Eimeria sp. given the presence of 4 sporocysts observed within the oocysts.
Species description
Family Eimeriidae Minchin, 1903
Genus Eimeria Schneider, 1875
Eimeria haematopusi n. sp.
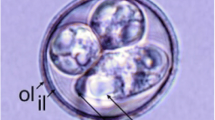
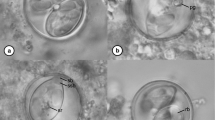
Oocysts (n = 20) ellipsoidal, 20–21 × 12–13 (20.1 × 12.9) μm; length/width (L/W) ratio 1.5–1.6 (1.56). Wall bi-layered, 0.7–1.2 (0.9) μm thick, outer layer smooth to slightly rough, c.2/3 of total thickness (Table 1.). Micropyle present, robust, and protruding, 3.5–4.3 (3.9) μm wide; occasionally with a rounded polar body attached to the micropyle. Oocyst residuum presents as a compact irregular mass, in addition to two to five polar granules being present. Sporocysts (n = 20) ellipsoidal, 9–11 × 5–6 (10.1 × 5.9) μm; L/W ratio 1.6–2.1 (1.71). Stieda body present, flattened to half-moon-shaped, 0.3–0.4 × 1.2–1.3 (0.4 × 1.3) μm; sub-Stieda body and para-Stieda body absent; sporocyst residuum present, composed of a few spherules scattered among the sporozoites. Sporozoites with anterior and posterior refractile bodies, but the nucleus is indiscernible (Fig. 1 & 2).
Photomicrographs of sporulated oocysts of E. haematopusi n. sp. from the Australian pied oystercatcher Haematopus longirostris. Note the inner layer (il) and smooth/rough outer layer (sol/rol) in the oocyst wall, micropyle (m), oocyst residuum (or), polar granule (pg), refractile body (rb) and Stieda body (sb). Scale bar: 10 μm
Type-host: Australian pied oystercatcher (Haematopus longirostris Vieillot) (Aves: Charadriiformes: Haematopodidae)
Other hosts: Unknown.
Type-locality: Heirisson Island (− 31° 57′ 35.39″ S, 115° 52′ 34.19″ E), Perth, Western Australia, Australia.
Type-material: Oocysts in 10% formalin and oocyst photo-syntypes were deposited in the Western Australian Museum under the reference number WAM Z68805.
Prevalence: 1/1 (100%).
Prepatent period: Unknown.
Patent period: Unknown.
Site of infection: Unknown, oocysts collected from faeces.
Sporulation time: 48–72 hours.
Oocysts in 10% formalin and oocyst photosyntypes were deposited in the Western Australian Museum under the reference number WAM Z68803.
Representative DNA sequences: The newly generated sequences are deposited in the GenBank database under the accession numbers OR568560 and OR568561 for 18S rRNA Clone 1 and Clone 2, respectively.
ZooBank registration: To comply with the regulations set out in Article 8.5 of the amended 2012 version of the International Code of Zoological Nomenclature (ICZN, 2012) details of the new species have been submitted to ZooBank. The Life Science Identifier LSIDurn:lsid:zoobank.org:pub:2366D36B-A3C1-49A0-A330-FE958E0448B6. The LSID for the new name Eimeria haematopusi is LSIDurn:lsid:zoobank.org:act:5960EF64-64E7-4DA0-94B5-934B8801CC37.
Phylogenetic analysis of 18S rRNA gene
A total of eight positive colonies containing the pGEMT- E. haematopusi18S rRNA vector were picked up and sequenced in both directions. Two types of unique sequences were obtained and were named Clone1 and Clone 2, at 1286 and 1289 bp, respectively. The two types of 18S rRNA sequences were aligned with 19 Eimeria spp., five Isospora spp. and one Caryospora sp. sequences isolated from birds. The justification for the selection of the reference sequences chosen was based on the NCBI Blast similarities (one sequence per species) and included sequences from Eimeria spp. relevant to this study. A sequence of Toxoplasma gondii (Nicolle and Manceaux) (EF472967) was used as the outgroup. The E. haematopusi n. sp. sequences from Clone 1 and 2 shared the highest genetic similarity of 97.6% and 97.2%, respectively with E. reichenowi (AB544308), which was identified from a hooded crane (Grus monacha Temminck) in Japan. As shown in Fig. 3, E. haematopusi n. sp. Clone 1 and 2 grouped within a clade with Eimeria spp. isolated from wetland birds and include E. paludosa (KJ767187) from a dusky moorhen (Gallinula tenebrosa) in Western Australia, E. reichenowi (AB544308) and E. gruis (AB544336) both from hooded cranes (G. monacha) and Eimeria bosquei (MF503487) from a black-necked crane (Grus nigricollis Przhevalsky) in China.
Discussion
To our knowledge, there have been no previous reports of Eimeria species identified from the Australian pied oystercatcher (H. longirostris). This is the first description of Eimeria identified from an Australian oystercatcher.
Coccidia infections in birds may result in disease or may be subclinical. There are more than 197 Eimeria species infecting birds worldwide (Yabsley, 2008). Most of these complete their lifecycle in the alimentary tract. Enteric disease caused by pathogenic Eimeria species are responsible for large losses in the poultry industry worldwide. A small number of Eimeria species undergo extra intestinal development stages in the kidneys, for example Eimeria truncata in geese or throughout the body (disseminated visceral coccidiosis) as occurs with Eimeria reichenowi and Eimeria gruis in cranes (Samour, 2016). Both Eimeria and Isopora can commonly be found in the kidneys of wild geese, ducks, and shorebirds while hepatic coccidiosis due to Eimeria is seen in mutton birds and little penguins (Obendorf and McColl, 1980). In a previous study, E. haematopi was described from the kidneys of a Eurasian oystercatcher (H. ostralegus) during investigations of parasites in sea and coastal birds along the German North Sea coast (Gottschalk and Prange, 2011). In a similar study, coccidian infections were identified in two Eurasian oystercatchers (H. ostralegus) during histological examination of kidneys, during investigations of the health status of seabirds along the North Sea coast of Germany (Siebert et al., 2012). In the later study, the coccidia were not identified, limiting comparisons between Eimeria species isolated from H. ostralegus. Morphological and molecular characterization of the oocysts isolated from the pied oystercatcher in this study revealed that it was a novel species of Eimeria.
It would have been advantageous if histopathological studies had been performed on the bird in this study however the bird was sent for incineration before these studies could be conducted, It is also not possible to know if the coccidian infection played a role in the death of the bird as no necropsy was done and it may well have been an incidental finding.
Morphological comparison between the Eimeria species from the Australian and Eurasian oystercatchers showed oocysts of E. haematopusi n. sp. to be smaller in size and ellipsoid in shape compared to the oocysts of E. haematopi which are ovoid and larger. Unfortunately, no other morphological comparisons could be made between these two Eimeria species due to the limited features described for E. haematopi (Gottschalk and Prange, 2011) (http://eimeria.unl.edu/charad.html) (accessed on 20 Sep. 2023). When the morphological parameters of E. haematopusi n. sp. was compared to genetically similar Eimeria species E. reichenowi, E. paludosa and E. gruis no obvious similarities were identified. The oocysts of E. reichenowi, E. paludosa and E. gruis all measure smaller than E. haematopusi n. sp., have generally ovoid shaped sporocysts and present with a sub-Stieda body residuum. This is in contrast with E. haematopusi n. sp., which is larger in size, ellipsoidal in shape and lacks a sub-Stieda body residuum.
Unfortunately, no genetic comparisons could be made between the two Eimeria species (E. haematopusi n. sp. and E. haematopi) isolated from oystercatchers to date, given the lack of genetic data for E. haematopi. Comparison between other Eimeria species however showed E. haematopusi n. sp. to be most closely related to E. reichenowi (AB544308), (genetic similarity of 97.6% and 97.2% from Clone 1 and 2, respectively) identified from a hooded crane (G. monacha) in Japan and less like Eimeria briceae from an Australian pelican (Pelecanus conspicillatus Temminck) with genetic similarities of 96.1% to Clone 1 and 95.6% to Clone 2. The close association of E. haematopusi n. sp. to Eimeria species isolated from a variety of different host species including the hooded crane (G. monacha) and dusky moorhen (G. tenebrosa) suggest that the evolution of this parasite does not solely occur along with their hosts but that the different ecological environments are also important in this long evolutionary process. Genetic comparisons between Eimeria isolates from different species of oystercatchers would be vital for such clarification.
In this study, we describe a novel species of Eimeria from the Australian pied oystercatcher and contribute to the first genetic characterisation of Eimeria species described from the Haematopodidae family. To understand the health impacts of this parasite, further studies are needed to ascertain the effects this Eimeria species has on its host.
Conclusion
Morphological and genetic comparison of E. haematopusi n. sp. with other known Eimeria spp. revealed that the coccidian identified in the wild Australian pied oystercatcher is a new species. E. haematopusi n. sp. is the first coccidian to be reported from an Australian pied oystercatcher and is the first sequence available of a coccidian from this bird species. The morphological and genetic sequence information from our study of E. haematopusi n. sp. further contributes towards our knowledge of Eimeria spp.
Data availability
No datasets were generated or analysed during the current study.
References
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403–410.
Benson, D. A., Karsch-Mizrachi, I., Lipman, D. J., Ostell, J., & Wheeler, D. L. (2005). GenBank. Nucleic Acids Research, 33 (Database issue), D34–D38. https://doi.org/10.1093/nar/gki063
Berto, B. P., Flausino, W., McIntosh, D., Teixeira-Filho, W. L., & Lopes, C. W. (2011). Coccidia of New World passerine birds (Aves: Passeriformes): A review of Eimeria Schneider, 1875 and Isospora Schneider, 1881 (Apicomplexa: Eimeriidae). Systematic Parasitology, 80, 159–204. https://doi.org/10.1007/s11230-011-9317-8
Berto, B. P., Machado, E. L., Hossotani, C. M. S., Beretta B. M. S., da Silva D. R. R., Nakamura, A. A., & Meireles, M. V. (2023). Integrative taxonomy for the traditional coccidians (Chromista: Miozoa: Eimeriidae) from island canaries (Aves: Passeriformes: Fringillidae): Worldwide distribution, morphological and molecular characterization, revaluations and establishment of junior synonyms. Systematic Parasitology, 100, 245–259. https://doi.org/10.1007/s11230-023-10084-6
Coker, S. M., McInnes, K., Vallee, E., Biggs, P., Pomroy, W. E., Howe, L., & Morgan, K. J. (2023). Molecular characterisation and additional morphological descriptions of Eimeria spp. (Apicomplexa: Eimeriidae) from brown kiwi (Apteryx mantelli Bartlett). Systematic Parasitology, 100, 269–281. https://doi.org/10.1007/s11230-023-10086-4
Courtney, C. H., Forrester, D. J., Ernst, J. V., & Nesbitt, S. A. (1975). Coccidia of sandhill cranes, Grus canadensis. Journal of Parasitology, 61, 695–699.
Duszynski, D. W. (2021). Biodiversity of the Coccidia (Apicomplexa: Conoidasida) in vertebrates: What we know, what we do not know, and what needs to be done. Folia Parasitol (Praha). https://doi.org/10.14411/fp.2021.001
Duszynski, D. W., Couch, L., & Upton, S. (2001). The coccidia of Charadriiformes (auks, avocets, curlews, gulls, ibisbills, jacanas, lapwings, murres, oystercatchers, plovers, pratincoles, puffins, sandpipers, seedsnipes, sheathbills, skimmers, skua, snipe, stilts, terns, turnstones, woodcock, yellow legs). NSF Grant PEET DEB 9521687. https://www.k-state.edu/parasitology/worldcoccidia/CHARADRIIFORMES
Gottschalk, C., & Prange, H. (2011). Parasites in birds at Mellum, an island in German coastal waters of the North Sea (in German). Der Zoologische Garten N. F., 80, 29–86.
Hockey, P., Kirwan, G. M., & Boesman, P. F. D. (2020). Pied Oystercatcher (Haematopus longirostris), version 1.0. In J. del Hoyo, A. Elliott, J. Sargatal, D. A. Christie, & E. de Juana (Eds.), Birds of the world. Cornell Lab of Ornithology. https://doi.org/10.2173/bow.pieoys1.01
International Commission On Zoological Nomenclature. (2012). Amendment of Articles 8, 9, 10, 21 and 78 of the International Code of Zoological Nomenclature to expand and refine methods of publication. Zookeys, 219, 1–10.
Jeanes, C., Vaughan-Higgins, R., Green, R. E., Sainsbury, A. W., Marshall, R. N., & Blake, D. P. (2013). Two new Eimeria species parasitic in corncrakes (Crex crex) (Gruiformes: Rallidae) in the United Kingdom. Journal of Parasitology, 99, 634–638.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Molecular Biology and Evolution, 35, 1547–1549.
Lainson, R. (1994). Observations on some avian coccidia (Apicomplexa: Eimeriidae) in Amazonian Brazil. Memórias do Instituto Oswaldo Cruz, Rio de Janeiro, 89, 303–311.
Mandal, A. K. (1965). Studies on some aspects of avian coccidia [Protozoa: Sporozoa] 3. Five new species of the genus Eimeria Schneider, and a new subspecies of Eimeria roscoviensis (Labbe). Proceedings of the Zoological Society Calcutta, 18, 47–57.
McAllister, C. T., & Upton, S. J. (1990). Description of the oocysts of Eimeria paludosa (Apicomplexa: Eimeriidae) from Fulica americana (Aves: Gruiformes), with comments on synonyms of eimerian species from related birds. Journal of Parasitology, 76, 27–29.
Medina, J. P., Medina-Valdez, H., Sánchez-Jasso, J. M., García-Albarrán, M., Salgado-Miranda, C., & Soriano-Vargas, E. (2019). Eimeria aegoliusia n. sp. (Sporozoa: Eimeriidae) from the northern saw-whet owl Aegolius acadicus (Gmelin) (Strigiformes: Strigidae) in Mexico. Systematic Parasitology, 96, 521–526. https://doi.org/10.1007/s11230-019-09863-x
Obendorf, D., & McColl, K. (1980). Mortality in little penguins (Eudyptula minor) along the coast of Victoria. Journal of Wildlife Diseases, 16, 251–259.
Parker, B. B., & Duszynski, D. W. (1986). Coccidiosis of Sandhill Cranes (Grus canadensis) wintering in New Mexico. Journal of Wildlife Diseases, 22, 25–35.
Pizzey, G., & Knight, F. (2007). The field guide to the birds of Australia (p. 474) (P. Menkhorst, Ed.). Harper Collins Publishers Pty Limited.
Samour, J. (Ed.). (2016). Avian medicine (3rd ed.). Elsevier.
Siebert, U., Schwemmer, P., Guse, N., Harder, T., Garthe, S., Prenger-Berninghoff, E., & Wohlsein, P. (2012). Health status of seabirds and coastal birds found at the German North Sea coast. Acta Veterinaria Scandinavica, 54, Article Number 43.
Winkler, D. W., Billerman, S. M., & Lovette, I. J. (2020). Oystercatchers (Haematopodidae), version 1.0. In S. M. Billerman, B. K. Keeney, P. G. Rodewald, & T. S. Schulenberg (Eds.), Birds of the world. Cornell Lab of Ornithology. https://doi.org/10.2173/bow.haemat1.01
Woodyard, E. T., Rush, S. A., & Rosser, T. G. (2019). Redescription of Eimeria megabubonis Upton, Campbell, Weigel & McKown, 1990 (Apicomplexa: Emeriidae) from the great horned owl Bubo virginianus (Gmelin). Systematic Parasitology, 96, 585–594. https://doi.org/10.1007/s11230-019-09867-7
Yabsley, M. J. (2008). Eimeria. In C. T. Atkinson, N. J. Thomas, & D. B. Hunter (Eds.), Parasitic diseases of wild birds. Wiley-Blackwell.
Yang, R., Brice, B., Al Habsi, K., Elliot, A., & Ryan, U. (2015). Isospora streperae n. sp. (Apicomplexa: Eimeriidae) from a grey currawong (Strepera versicolor plumbea) (Passeriformes: Artamidae) in Western Australia. Experimental Parasitology, 151–152, 49–55.
Yang, R., Brice, B., Berto, B. P., & Ryan, U. (2021). Morphological and molecular description of a new species of Isospora (Apicomplexa) from a New Holland honeyeater (Phylidonyris novaehollandiae). Parasitology International, 83, 102348.
Yang, R., Brice, B., Elloit, A., Lee, E., & Ryan, U. (2014). Morphological and molecular characterization of Eimeria paludosa coccidian parasite (Apicomplexa: Eimeriidae) in a dusky moorhen (Gallinula tenebrosa, Gould, 1846) in Australia. Experimental Parasitology, 147, 16–22.
Yang, R., Brice, B., & Ryan, U. (2016). Morphological and molecular characterization of Isospora neochmiae n. sp. in a captive-bred red-browed finch (Neochmia temporalis) (Latham, 1802). Experimental Parasitology, 166, 181–188.
Yang, R., Brice, B., Ryan, U., & Bennett, M. D. (2013a). Eimeria tiliquae n. sp. (Apicomplexa: Eimeriidae) from the shingleback skink (Tiliqua rugosa rugosa). Experimental Parasitology, 133(2), 144–149.
Yang, R., Murphy, C., Song, Y., Ng-Hublin, J., Estcourt, A., Hijjawi, N., Chalmers, R., Hadfield, S., Bath, A., Gordon, C., & Ryan, U. (2013b). Specific and quantitative detection and identification of Cryptosporidium hominis and C. parvum in clinical and environmental samples. Experimental Parasitology, 135(1), 142–147.
Acknowledgements
The authors wish to thank Yasmin Hunter (former KWRC volunteer and wildlife rehabilitator), Western Australian Seabird Rescue (WASR) and all Kanyana Wildlife Rehabilitation Centre volunteers for their commitment and dedication to caring for wildlife. We are also grateful to the staff at both the Wattle Grove and Kalamunda Veterinary Hospitals for their specialist treatment and care of the wildlife treated at their clinics.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions.
Author information
Authors and Affiliations
Contributions
Conceptualization, B.B. and R.Y.; methodology, D.L. and A.Z.A.; investigation, J.A. and B.B; validation, H.G., B.B. and R.Y.; formal analysis, B.P.B.; A.E. and J.A; resources, B.B.; data curation, D.L.; writing-original draft preparation, J.A.; writing-review and editing, B.B., H.G., A.Z.A and R.Y; visualization, A.E.; supervision, R.Y.; project administration, B.B. and R.Y.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Austen, J.M., Brice, B., Liu, D. et al. Morphological and molecular characterization of Eimeria haematopusi n. sp. (Apicomplexa: Eimeriidae) in an Australian Pied Oystercatcher (Haematopus longirostris) (Aves: Charadriiformes). Syst Parasitol 101, 40 (2024). https://doi.org/10.1007/s11230-024-10152-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11230-024-10152-5