Abstract
Serpentirhabdias mexicanus n. sp. (Nematoda: Rhabdiasidae) is described from the lung of the nauyaca viper Bothrops asper in Puebla State, central Mexico. This new species is the fifth of the genus described having onchia. Among the species included in this group, the new species is morphologically closest to S. viperidicus and S. atroxi. However, it differs from both species mainly by having only one excretory gland (compared to two present in S. viperidicus and S. atroxi). In addition, S. mexicanus n. sp. can be separated of S. viperidicus by tail length, shape of vulval lips, geographic distribution and host species and from S. atroxi by body length, number of papillae in the cephalic region, as well as the host species and geographic distribution. In the present study, we propose the new species based on morphological, host spectrum and genetic evidence. Phylogenetic analysis indicated Serpentirhabdias as a monophyletic group, with two subgroups that are congruent with the presence/absence of onchia in the esophagostome, host association and other relevant morphological characters.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Rhabdiasidae Railliet, 1915, a family of nematodes parasitic mainly of lungs of amphibian and reptiles, comprises eight genera: Acanthorhabdias Pereira, 1927, Chabirenia Lhermitte-Vallarino, Bain, Deharo, Bertani, Voza, Attout & Gaucher, 2005, Entomelas Travassos, 1930, Kurilonema Szczerbak & Sharpilo, 1969, Neoentomelas Hasegawa, 1989, Pneumonema Johnston, 1916, Rhabdias Stiles & Hassall, 1905 and Serpentirhabdias Tkach, Kuzmin & Snyder, 2014 (Tkach et al., 2014). Currently, Serpentirhabdias includes twenty valid species (Kuzmin and Tkach, 2022). Eleven of these have been registered in the Americas, parasitising members of Serpentes; particularly from Viperidae, only two species are known: S. viperidicus Morais et al., 2017 and S. atroxi Kuzmin et al., 2016 recorded in the Brazilian Neotropics (Kuzmin et al., 2016; Morais et al., 2017). In Mexico, two species have been reported infecting colubrid snakes: S. lamothei (Martínez-Salazar and León-Règagnon, 2006) parasite of Leptodeira maculata (Hallowel) in the States of Jalisco, Colima and Michoacán, on the Pacific slope of this country, and S. fuscovenosa (Railliet, 1899) in the States of Campeche, Guerrero, Jalisco, Michoacán, Oaxaca, Puebla, Querétaro, Veracruz, and Yucatán (Martínez-Salazar and León-Règagnon, 2006). During an ongoing project to increase the inventory of the helminth fauna associated to Mexican reptiles, we found several specimens of an undescribed species of Serpentirhabdias parasitising the nauyaca viper Bothrops asper (Garman) (Viperidae), which is described herein based on morphometric and molecular evidence.
The nauyaca viper is a poisonous snake with wide distribution from sea level up to 1,300 meters above sea level (masl), inhabiting tropical and subtropical forests of Mexico in the States of Querétaro, Hidalgo, San Luis Potosí, Tamaulipas, Puebla, Veracruz, Campeche, Tabasco, Quintana Roo, Oaxaca, Chiapas and Yucatán (Campbell et al., 2004; Saldarriaga-Cordoba et al., 2017). The snake has large body size reaching 1.50 to 2.0 meters. The new species of lungworm described herein is the first recognized species of Serpentirhabdias parasitising a member of Viperidae from Mexico and North America.
Materials and Methods
A female of B. asper was collected in Tlatlauquitepec, Puebla, Mexico, near La Soledad Dam (19° 57′ 59.1″ N, 97° 26′ 50.9″ W; 796 masl) in August 2022. Total length (TTL; 1.34 m), snout-vent length (SVL; 1.17 m) and tail length (TL; 0.17 m) was obtained using a measuring tape (accuracy: 0.01 m); body weight (BW; 588.2g) was measured with a digital scale. The snake was found dead with a 10 cm fresh wound behind the head, inflicted with a bowie knife by local people. Host was collected under permission issued by the Secretaría de Medio Ambiente y Recursos Naturales (SGPA/DGVS/03184/22). The specimen was put inside a cooler filled with ice and transferred to the Laboratorio de Helmintología, Instituto de Biología-UNAM (IBUNAM). In the laboratory, it was dissected in order to recover the helminths following Lamothe-Argumedo (1997) and subsequently deposited in the Colección Nacional de Anfibios y Reptiles (CNAR) at IBUNAM (IBH 35835).
Lungworms were removed from the lung, counted in situ and placed in saline (0.65%). For their morphological study, some worms were fixed in 4% hot formalin and preserved in ethanol 70%; the remaining specimens were placed directly in absolute ethanol and stored at −4 °C for molecular procedures. Lungworms were cleared with 1:1 mixture of alcohol-glycerin, as a temporary mounting medium for examination under light microscopy. Measurements (mean followed by range and standard deviation) are in micrometers if no other unit is indicated. Line drawings were made with a microscope equipped with a camera lucida. Identification of worms required observation under a scanning electron microscope (SEM); therefore, some nematodes were dehydrated in a graded ethanol series and dried to critical point with CO2, then coated with a gold–palladium mixture and mounted on metal stubs with silver paste. Specimens were studied with a Hitachi Stereoscan Model S2469N at 15 kV at Laboratorio Nacional de Biodiversidad (LANABIO), IBUNAM. Type specimens were deposited at Colección Nacional de Helmintos (CNHE), IBUNAM.
DNA extraction, PCR amplification and sequencing
Five nematodes previously fixed in absolute ethanol were randomly selected and put separately in 1.5 mL microcentrifuge tubes at room temperature, waiting for evaporation of the ethanol excess to be eliminated. Tissue digestion and DNA extraction were performed with EZ-10 Spin Column Genomic DNA Minipreps Kit, Bio Basic Inc. (Ontario, Canada) according to the manufacturer's instructions.
Mitochondrial cytochrome c oxidase subunit 1 (COI) was amplified. PCR reactions were prepared in a total volume of 15 μL containing: 2 μL of template DNA, 0.2 μL of each primer (LCO1490: 5′-GGTCAACAAATCATAAAGATATTGG -3′ and HCO2198: 5′-TAAACTTCAGGGTGACCAAAAAATCA-3′) (Folmer et al., 1994), 3 μL of 5x MyTaq Reaction Buffer and 0.1 μL MyTaq DNA Polymerase (Bioline Cat. BIO-21105), and 9.5 μL of RNAse-free H2O. Amplification started with an initial denaturing of 94 °C for 5 min, then 30 cycles with a first step of 94 °C for 45 s, a second step of 48 °C for 45 s and a third step of 72 °C for 1 min, and a final extension of 72 °C for 7 min. PCR reaction products were visualized by agarose gel electrophoresis and purified using CentriSep 96 filter plates (ThermoFisher Scientific) with Sephadex G-50 (Cytiva, Marlborough, Massachusetts). Sequencing reactions were prepared in a total volume of 10 μL using 0.4 μL of BigDye Terminator v. 3.1 (Applied Biosystems®, Waltham, Massachusetts), 2 μL of 5× Reaction Buffer, 4 μL of ddH2O, 1 μL of primer [10 μM], and 3 μL of purified PCR product. Samples were purified using Sephadex G-50 then 25 μL of 0.5 mM EDTA was added to each sample to be finally sequenced in an ABI-PRISM 3100 (Applied Biosystems®, Waltham, Massachusetts) sequencer instrument at LANABIO, IBUNAM.
Phylogenetic analysis
For the phylogenetic analysis, we used as outgroup some sequences of Rhabdias species available in GenBank following Müller et al. (2018) (Table 1); we constructed a matrix with a total of 19 COI gene sequences; to align those sequences we used the online version of MAFFT v.7 (Katoh et al., 2019) with default parameters and made a final manual editing of the endpoints in Mesquite v. 3.51 (Maddison and Maddison, 2018). JModeltest v. 3.0 was used to infer the best evolution model (Anderson and Burnham, 2004). Phylogenetic analysis was performed with Maximum Likelihood (Stamatakis et al., 2005), bootstrap-based resampling with 10,000 iterations to assess nodal supports. Bayesian inference was performed with the program MrBayes v. 3.2.7 (Ronquist et al., 2012). The settings were fixed as follows: 2 simultaneous runs with 4 Markov Chains Monte Carlo (MCMC) for 10 million generations, sampling every 1000 generations, a heating parameter value of 0.2, and a “burn-in” of 25%. The convergence statistics were checked using Tracer v. 1.7 (Rambaut et al., 2018). A 50% majority-rule consensus tree representing the posterior probability distribution of clades was produced for the sampled trees. Trees were visualized in Fig Tree v.1.4.4 (Rambaut, 2006). Additionally, uncorrected pairwise p-distances were calculated using the program MEGAX and converted to percentage difference by multiplying the p- distance value by 100 (Stecher et al., 2020).
Results
Serpentirhabdias mexicanus n. sp. Velázquez-Brito, García-Prieto, León-Règagnon & Garduño-Montes de Oca
Phylum Nematoda Cobb, 1932
Class Chromadorea Inglis, 1983
Order Rhabditida Chitwood, 1933
Family Rhabdiasidae Railliet, 1916
Genus Serpentirhabdias Tkach, Kuzmin & Snyder, 2014
Taxonomic summary
Type host: Nauyaca or terciopelo pit viper Bothrops asper (Garman) (Serpentes: Viperidae).
Type locality: Tlatlauquitepec, Puebla, México (19° 57′ 59.1″ N; 97° 26′ 50.9″ W).
Site of infection: Lung.
Prevalence: 100% (1 infected host out of 1 examined).
Intensity of infection: 29 individuals/host.
Material deposited: Holotype, female, Colección Nacional de Helmintos del Instituto de Biología, Universidad Nacional Autónoma de México, CNHE 11875; paratypes CNHE 11876 and 11877.
GenBank accession: COI sequences OR583874, OR883410, OR883411, OR883412 and OR883413.
Etymology: The new species is named after the country in which it was found.
Morphological description
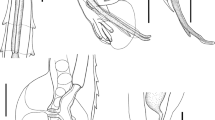
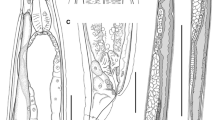
Based on ten adult females. Body 5.88 (5.247–6.361 ± 0.306) mm long. Cuticle slightly inflated at anterior [6.25 (1–13 ± 3.575) wide] and posterior ends of body (Figs. 1A, 1B, 1C, 2A and 2B). Anterior end rounded (Figs. 1D, 2C); sharp and conical posterior end (Fig. 1C). Body width 280 (227–372 ± 42.66) at vulva level; 171 (136–236 ± 35.87) at esophageal–intestinal junction. Oral opening round and wide in apical view; with six lips similar in shape and size, arranged in two lateral groups of three each; each lip with sclerotized triangular small papillae (Figs. 1D, 2C, 3A). Buccal capsule and vestibulum absent. Three pairs of small and slightly sclerotized onchia in esophagostome (Fig. 2B). Club-shaped esophagus 302 (273–330 ± 19.99) long, representing 5.15% (4.29–5.73 ± 0.47) body length; ending in a dilated bulb 79 (73–86 ± 3.80) long by 75 (64–86 ± 7.80) wide. Nerve-ring encircling esophagus, posterior to its mid-length, 168 (159–175 ± 4.94) from anterior end of body (Figs. 1A, B and 2A). Intestine brownish-black in uncleared specimens, parallel to uterus, without loops along body, running towards rectum. Excretory pore small, posterior to nerve ring, at 203 (190–215 ± 7.61) from anterior end (Figs. 1A, B, and 2D); excretory duct thin and short. Sub-ventral excretory gland relatively thin and elongated; 0.429 (0.360–0.495 ± 0.06) long by 0.045 (0.036–0.054 ± 0.007) wide, located near to esophageal bulb, ending after the esophagus-intestinal junction (Figs. 1B, 2A). Reproductive system typical of Rhabdiasidae (amphidelphic). Vulva pre-equatorial, located at 2.65 (2.33–2.99 ± 0.230) mm from anterior end, papillae absent. Vulval lips slightly protruded (Fig. 2E). Uterus with numerous eggs [137 (120–150 ± 12.58)], in different stages of development; eggs located near vulva, fully developed with visible larvae. Eggs 74 (68–86 ± 6.58) × 37 (35–44 ±2.71) (Fig. 2G). Short rectum, slightly cuticularized (Figs. 1C, 2F). Conical and pointed tail (Figs. 1C and 2F). Tail length 153 (122–170 ± 12.75), representing 2.60% (2.13–2.99 ± 0.24) of total body length.
Serpentirhabdias mexicanus n. sp., light micrographs. (A) Anterior end, [nr] nerve ring, [e] esophagus, [eg] excretory gland, [in] intestine. (B) [on] Onchia lateral view. (C) Apical view. (D) [ep] Excretory pore. (D) [v] Vulvar protruding lips. (F) [t] Tail, lateral view. (G) [lg] Larvated egg. Scale Bar 20 µm (C, B, D, E, G), 50 µm (A, F).
Remarks
Serpentirhabdias mexicanus n. sp. was assigned to this genus due to the presence of six lips similar in shape and size, arranged in two lateral groups; body cuticle thin and without conspicuous inflation, vulva pre-equatorial and buccal capsule absent, as well as by inhabiting the lung of snakes (Tkach et al., 2014; Kuzmin et al., 2016).
Serpentirhabdias comprises twenty species distributed worldwide (Kuzmin and Tkach, 2022); species of the genus can be separated in two groups because of the presence (four species) or absence (sixteen species) of onchia in the esophagostome. The new species belongs to the group with onchia, together with S. moi Machado et al., 2018, S. mussuranae Kuzmin et al., 2020, S. atroxi Kuzmin et al., 2016 and S. viperidicus Morais et al., 2017. These four species also lack buccal capsule as in the new species. An interesting finding among the five species having onchia is the formation of two subgroups based on the morphology of the oral opening. Species that parasitize colubrids have a triangular oral opening (S. moi and S. mussuranae), while those that parasitize viperids have a rounded mouth opening (S. atroxi, S. viperidicus and S. mexicanus n. sp.).
Serpentirhabdias mexicanus n. sp. differs from S. viperidicus by the absence of a short vestibulum (present in S. viperidicus), the length of the tail [153 (122–170) and 201 (123–208), respectively), as well as by the presence of slightly protruded vulval lips in S. mexicanus n. sp. and not protruded vulval lips in S. viperidicus. Furthermore, the presence of two excretory glands in S. viperidicus (and only one in the new species) allows the two species to be differentiated. Other traits distinguishing these two species are geographic distribution and the type host: S. mexicanus n. sp. was found in B. asper in central Mexico, specifically in Puebla State, while S. viperidicus parasitizes B. moojeni in Brazil (Morais et al., 2017).
Serpentirhabdias mexicanus n. sp. can be distinguished from S. atroxi by body length [5.38 (5.25–6.36)] mm in the new species and [4.32 (3.4–4.5) mm in S. atroxi], but particularly by the number of excretory glands (1 in the new species and 2 in S. atroxi). In addition, S. atroxi presents small internal and external labial papillae, while S. mexicanus n. sp. only has one external papilla on each lip. From an ecological point of view, these species differ by their geographic distribution and type host; S. atroxi has been found in Brazil, parasitising B. atrox (Kuzmin et al., 2016).
Phylogenetic analysis
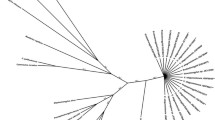
In the phylogenetic hypotheses obtained based on COI sequences, using either inference method, Serpentirhabdias is confirmed as a monophyletic genus; sequences of individuals of the new species are grouped in a well supported monophyletic clade. Species of Serpentirhabdias with onchia group together, and within this group, S. mexicanus n. sp. is phylogenetically closest to S. viperidicus, while S. mussuranae is related to S. moi (Fig. 4). According to the percentage genetic distance matrix presented in Table 2, the new species differs from the rest of its congeners in an interval that ranges between 3% (S. atroxi) and 16% (S. elaphe). Genetic divergence among sequences of 5 specimens of S. mexicanus was 0%.
Discussion
Serpentirhabdias mexicanus n. sp. is the third species of the genus recorded in Mexico along with S. lamothei and S. fuscovenosa (see Martínez-Salazar and León-Règagnon, 2006). However, it is the first species with onchia described in the country (see Kuzmin et al., 2020). Among Serpentirhabdias species, onchia were first described in S. atroxi. However, these structures have also been described in species of Entomelas and Rhabdias, and according to Kuzmin et al. (2016), this trait represents an evolutive convergence for these three genera.
Phylogenetic relationships within Rhabdiasidae are not well resolved, mainly due to the lack of molecular information. However, the clades recovered in the present analysis with COI sequences available in Genbank to date (Fig. 4), corroborated the monophyly of Serpentirhabdias as proposed by Morais et al. (2017), based on a phylogenetic analysis of ribosomal DNA. The present phylogenetic analysis of COI sequences, group species of Serpentirhabdias with onchia in a monophyletic clade, although Bayesian support was low. On the other hand, the assemblage of species according to the shape of oral opening and host family [round in species parasitising Viperidae (S. atroxi, S. viperidicus and S. mexicanus n. sp.)] and triangular in Colubridae (S. moi and S. mussuranae), was not supported. We consider that more solid conclusions on the relationships between the morphology of oral opening and host family of Serpentirhabdias spp. (Viperidae-Colubridae) will require a broader sampling effort, including wide geographic and host spectrum. It is worth noting that species with onchia have been described solely in the American continent, specifically in Brazil and Mexico (Kuzmin et al., 2016, 2021; Morais et al., 2017; Machado et al., 2018; present study). Erection of Serpentirhabdias mexicanus n. sp. as a new species is supported by morphological, molecular, ecological (geographic distribution and host) and phylogenetic evidence.
Data availability
The sequences generated and analyzed in this study have been deposited in the GenBank database under the accession numbers: OR583874, OR883410-13.
Consent for publication
Not applicable.
References
Anderson, D., and Burnham, K. (2004). Model selection and multi-model inference: A Practical Information-Theoretic Approach. Springer.
Campbell, J.A., Lamar, W.W., and Brodie, E.D. (2004). The venomous reptiles of the western hemisphere. Vol. 1. Cornell University Press.
Folmer O., Black M., Hoeh W., Lutz R., and Vrijenhoek R. (1994). DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology, 3, 294–299.
Katoh, K., Rozewicki, J., and Yamada, K.D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Briefing in Bioinformatics, 20, 1160–1166. https://doi.org/10.1093/bib/bbx108
Kuzmin, Y., du Preez, L., and Svitin, R. (2021). A new species of Serpentirhabdias Tkach, Kuzmin et Snyder, 2014 (Nematoda: Rhabdiasidae) parasitic in the herald snake, Crotaphopeltis hotamboeia (Laurenti) (Reptilia: Serpentes: Colubridae) in South Africa. Systematic Parasitology, 98, 477–485. https://doi.org/10.1007/s11230-021-09990-4
Kuzmin, Y., Giese, E.G., Melo, F.T.D.V., da Costa, P.A.F.B., Maschio, G.F., and Santos, J.N. D. (2016). Description of Serpentirhabdias atroxi n. sp. (Nematoda: Rhabdiasidae), a parasite of Bothrops atrox (Linnaeus) (Reptilia: Serpentes: Viperidae) in Brazilian Amazonia. Systematic Parasitology, 93, 37–45. https://doi.org/10.1007/s11230-015-9603-y
Kuzmin, Y., Halajian, A., Tavakol, S., Luus-Powell, W.J. & Tkach, V.V. (2017). Description and phylogenetic position of a new species of Rhabdias Stiles et Hassall, 1905 (Nematoda: Rhabdiasidae) from the banded rubber frog, Phrynomantis bifasciatus (Smith) (Amphibia: Microhylidae) in South Africa. Folia Parasitologica, 64, 035. https://doi.org/10.14411/fp.2017.035
Kuzmin, Y., and Tkach, V.V. (2022, October 2022). Rhabdias. Retrieved October 19, 2022, from http://izan.kiev.ua/ppages/rhabdias/list.htm.
Kuzmin, Y., Tkach, V.V., and Melo, F.T.D.V. (2020). Description, molecular characterization and life cycle of Serpentirhabdias mussuranae n. sp. (Nematoda: Rhabdiasidae) from Clelia clelia (Reptilia: Colubroidea) in Brazil. Journal of Helminthology, 94: e55. https://doi.org/10.1017/S0022149X19000348
Lamothe-Argumedo, R. (1997). Manual de técnicas para preparar y estudiar los parásitos de animales silvestres. AGT Editores.
Machado, S.A., Kuzmin, Y., Tkach, V.V., Dos Santos, J.N., Gonçalves, E.C., and Melo, F.T.D.V. (2018). Description, biology and molecular characterization of Serpentirhabdias moi n. sp. (Nematoda: Rhabdiasidae) from Chironius exoletus (Serpentes: Colubridae) in Brazil. Parasitology International, 6, 829–837. https://doi.org/10.1016/j.parint.2018.05.004
Maddison, W.P, and Maddison, D. R. (2018, April 2023). Mesquite: a modular system for evolutionary analysis. Version 3.51. Retrieved March 15, 2023, from http://www.mesquiteproject.org.
Martínez-Salazar, E. A., and León-Règagnon, V. (2006). Rhabdias lamothei n. sp. (Nematoda: Rhabdiasidae) from Leptodeira maculata (Colubridae) in Mexico, including new records of R. fuscovenosa (Railliet, 1899) Goodey, 1924. Zootaxa 1257, 27–48. https://doi.org/10.11646/zootaxa.1257.1.2
Morais, D.H., Aguiar, A., Müller, M.I., Narciso, R.B., Da Silva, L.A.F., and Da Silva, R.J. (2017). Morphometric and phylogenetic analyses of Serpentirhabdias viperidicus n. sp. (Nematoda: Rhabdiasidae) from the lance head snake Bothrops moojeni Hoge, 1966 (Reptilia: Serpentes: Viperidae) in Brazil. Journal of Helminthology, 91, 360–370. https://doi.org/10.1017/S0022149X16000377
Müller, M.I., Morais, D.H., Costa-Silva, G.J., Aguiar, A., Ávila, R.W., and Silva, R.J. (2018) Diversity in the genus Rhabdias (Nematoda, Rhabdiasidae): Evidence for cryptic speciation. Zoologica Scripta, 47, 595–607. https://doi.org/10.1111/zsc.12304
Müller, M.I., Morais, D.H., da Costa, L.F.S.T., de Vasconcelos Melo, F.T., Giese, E.G., Avila, R.W., and da Silva, R.J. (2023). Revisiting the taxonomy of Rhabdias fuelleborni Travassos, 1928 (Nematoda, Rhabdiasidae) with approaches to delimitation of species and notes on molecular phylogeny. Parasitology International, 92, 102692. https://doi.org/10.1016/j.parint.2022.102692
Prosser, S.W., Velarde-Aguilar, M.G., Leon-Règagnon, V., and Hebert, P.D. (2013). Advancing nematode barcoding: A primer cocktail for the cytochrome c oxidase subunit I gene from vertebrate parasitic nematodes Molecular Ecology Resources, 13, 1108–1115. https://doi.org/10.1111/1755-0998.12082
Rambaut, A. (2006, November, 2018). FigTree tree figure drawing tool (version 1.4. 2). Retrieved March 19, 2023, from http://tree.bio.ed.ac.uk/software/figtree/
Rambaut, A., Drummond, A.J., Xie, D., Baele, G., and Suchard, M.A. (2018). Posterior summarization in Bayesian Phylogenetics using Tracer 1.7. Systematic Biology, 67, 901–904. https://doi.org/10.1093/sysbio/syy032
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D.L., Darling, A., Höhna, S., and Huelsenbeck, J.P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. https://doi.org/10.1093/sysbio/sys029
Saldarriaga-Córdoba, M., Parkinson, C.L., Daza, J.M., Wüster, W., and Sasa, M. (2017). Phylogeography of the Central American lance head Bothrops asper (Serpentes: Viperidae). PLoS One, 12, e0187969. https://doi.org/10.1371/journal.pone.0187969
Stamatakis, A., Ludwig, T., and Meier, H. (2005). RAxML-III: a fast program for maximum likelihood-based inference of large phylogenetic trees. Bioinformatics, 21, 456–463. https://doi.org/10.1093/bioinformatics/bti191.
Stecher, G., Tamura, K., and Kumar, S. (2020). Molecular evolutionary genetics analysis (MEGA) for macOS. Molecular Biology and Evolution, 37, 1237–1239. https://doi.org/10.1093/molbev/msz312
Tkach, V.V., Kuzmin. T., and Snyder, S.D. (2014). Molecular insight into systematics, host associations, life cycles and geographic distribution of the nematode family Rhabdiasidae. International Journal of Parasitology, 44, 273–284. https://doi.org/10.1016/j.ijpara.2013.12.005
Willkens, Y., Rebelo, G.L., Santos, J.N., Furtado, A.P., Vilela, R.V., Tkach, V.V., Kuzmin, Y., and Melo, F.T.V. (2019). Rhabdias glaurungi sp. nov. (Nematoda: Rhabdiasidae), parasite of Scinax gr. ruber (Laurenti, 1768) (Anura: Hylidae), from the Brazilian Amazon. Journal of Helminthology, 94, E54. https://doi.org/10.1017/S0022149X1900047
Acknowledgments
We thank Laura Márquez, Nelly López, and Berenit Mendoza from Laboratorio Nacional de la Biodiversidad (LANABIO), for their help with sequencing of samples and the SEM pictures respectively. Special thanks to Georgina Ortega-Leite for providing important bibliographic references and César Hernández Hernández for his assistance in the collection of the nauyaca viper and Posgrado en Ciencias Biológicas, UNAM (CVU 696855, CV480725 and CVU 413071). Specimen was collected under the SEMARNAT Capture Permit (SGPA/DGVS/03184/22) granted to Alejandro Francisco Oceguera Figueroa.
Funding
This project was supported by the Programa de Apoyo a Proyectos de Investigación e Innovación Tecnológica (PAPIIT) IN 212119 to Fausto Roberto Méndez de la Cruz and Consejo Nacional de Ciencia y Tecnología (CONACYT) Project 220408 to VLR, and Posgrado en Ciencias Biológicas.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material collection, preparation, data collection and analysis were performed by AV-B, UG-MO, VMS-J and MCV-C. The first draft of the manuscript was written by LG-P and VL-R and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the author(s).
Ethical approval
Not applicable.
Informed consent
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Velázquez-Brito, A., García-Prieto, L., Garduño-Montes de Oca, U. et al. Serpentirhabdias mexicanus n. sp. (Nematoda: Rhabdiasidae), a parasitic lungworm of the nauyaca viper Bothrops asper (Serpentes: Viperidae) in the Mexican Neotropics. Syst Parasitol 101, 19 (2024). https://doi.org/10.1007/s11230-023-10144-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11230-023-10144-x