Abstract
Bunocotylid trematodes represent a group of 149 species with a rather complex taxonomic history. The current concept of the subfamily only includes three genera, Bunocotyle, Saturnius, and Robinia. Specimens of a bunocotylid were collected from the silver mullet, M. curema, from a coastal lagoon of Yucatán and identified as belonging to Saturnius. Further detailed morphological study revealed they corresponded to S. maurepasi, a species previously reported from the stripped mullet, Mugil cephalus in Mississippi, USA. Specimens were sequenced for the LSU of nuclear ribosomal RNA gene (28S) to test their phylogenetic position. We discovered that they do not belong in Saturnius since they nest as an independent lineage which is the sister taxa of a clade formed by Robinia, and Saturnius + Bunocotyle; additionally, the new genus exhibits high genetic divergence (10-12%) with respect to species allocated in the other bunocotylid genera. The species S. maurepasi was then transferred to the new genus as Parasaturnius maurepasi n. gen., n. comb. that was created to accommodate it, and was redescribed based on newly sampled specimens.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Members of Bunocotylidae Dollfus, 1950 are a relatively diverse group of trematodes with approximately 149 species infecting the gastrointestinal tract (mainly the stomach) of marine and estuarine fishes around the world (Atopkin et al., 2020; WoRMS, 2023). The taxonomic history and classification of the family has been rather controversial (Pankov et al., 2006; Sokolov et al., 2019; Louvard et al., 2022; Martin et al., 2023). Using a molecular phylogenetic analysis based on 28S rDNA, Atopkin et al. (2020) showed that Bunocotylidae was constituted by four subfamilies, namely Bunocotylinae Dollfus, 1950 with three genera, Hysterolecithinae Yamaguti, 1958 with four, Opisthadeninae Yamaguti, 1970 with five, and Quadrifoliovariinae Yamaguti, 1955 with three genera (see Worms, 2023). Nevertheless, in recent molecular phylogenetic analyses, these subfamilies of Bunocotylidae were yielded as paraphyletic (Louvard et al., 2022; Faltýnkova et al., 2022). In a recently published study, Martin et al. (2023) also suggested that Bunocotylidae was paraphyletic relative to the Hemiuridae sensu stricto and the Lecithasteridae sensu stricto. Based on the latter arguments, the current concept of Bunocotylinae contains only 14 species classified in three genera, Bunocotyle Odhner, 1928, with four species; Saturnius Manter, 1969, with nine; and Robinia Pankov, Webster, Blasco-Costa, Gibson, Littlewood, Balbuena & Kostadinova, 2006, with only one species (Pankov et al., 2006; Blasco-Costa et al., 2008; Louvard et al., 2022; Faltýnkova et al., 2022).
Members of this subfamily are mostly parasites of the stomach of mullets of the genus Mugil (L.) since 11 of the 14 species have been described in that fish group (Overstreet, 1977; Blasco-Costa et al., 2006; 2008; Marzoug et al., 2014). Only three species of bunocotylids have been described in the Americas, i.e., Bunocotyle sudatlantica Parukhin, 1976 from an unidentified fish belonging to Chaetodontidae Rafinesque, 1815 in Brazil; Saturnius maurepasi Overstreet, 1977 from the stripped mullet Mugil cephalus (L.) in Mississippi, USA; and Saturnius belizensis Fischtal, 1977 from the silver mullet Mugil curema Valenciennes in Belize (Kohn et al., 2007; Blasco-Costa et al., 2008; WoRMS, 2023). Overall, only four of the 14 species have been sequenced for some molecular marker, including Saturnius gibsoni Marzoug, Rima, Boutiba, Georgieva, Kostadinova and Pérez-del-Olmo, 2014; S. minutus Blasco-Costa, Pankov, Gibson, Balbuena, Raga, Sarabeev and Kostadinova, 2006; Bunocotyle progenetica (Markowski, 1936); and Robinia aurata Pankov, Webster, Blasco-Costa, Gibson, Littlewood, Balbuena and Kostadinova, 2006 (Pankov et al., 2006; Marzoug et al., 2014).
During a survey on the parasites of marine and estuarine fishes of the Yucatán Peninsula, specimens of the silver mullet, Mugil curema (Valenciennes) were collected in La Carbonera coastal lagoon, in Yucatán. Specimens of a bunocotylid trematode were obtained from the stomachs of their hosts. After comparing morphologically with the original descriptions of members of the family, we identified our specimens as Saturnius maurepasi, with a slight morphological variation. However, while conducting a molecular phylogenetic analysis of the large subunit of the ribosomal gene (28S rDNA), we unexpectedly discovered that our specimens formed a separate monophyletic clade from Saturnius spp., suggesting that they represented an undescribed genus. Here, we present the diagnosis of the genus Parasaturnius n. gen. to include S. maurepasi as a new combination, using morphological and molecular evidence, and we discuss the interrelationships within Bunocotylidae.
Materials and methods
Host collection and morphological analysis
A total of 10 individuals of M. curema (8–10 cm) were collected in May 2022 in La Carbonera coastal lagoon, Yucatán State, Mexico (21° 08′ 1.5′′ N, 90° 07′ 55.9′′ W) using cast nets; silver mullets were kept alive and examined for helminths a few hours after capture. Individual fish were euthanized by spinal severance (pithing) following the procedures accepted by the American Veterinary Medical Association (AVMA, 2020), dissected, and immediately examined under a stereomicroscope. Bunocotylids were recovered from the stomach lining of seven of the 10 host examined, fixed in hot distilled water, and preserved in 100% ethanol for morphological and molecular analyses.
Some unflatten specimens were post-fixed in hot formalin to harden the tegument. Specimens were dehydrated through graded alcohol series, stained with Mayer’s paracarmine (Merck, Darmstadt, Germany), cleared with methyl salicylate, and mounted on microscope slides with Canada balsam. Mounted specimens were examined under a bright field Nikon DS-Ri1 microscope. Measurements were taken using Nikon NIS Elements microscope software (Nikon) and are given in micrometres (μm). Drawings were made with Adobe Illustrator 25.4.1 (Adobe, Inc). Vouchers were deposited in the Colección Nacional de Helmintos (CNHE), Instituto de Biología, Universidad Nacional Autónoma de México, Mexico City.
For scanning electron microscopy (SEM), specimens were dehydrated in a graded ethanol series, critical point dried, and sputter coated with gold. Then, specimens were examined with a Hitachi Stereoscan Model SU1510 scanning electron microscope at 15 kV at the Laboratorio de Microscopia y Fotografía de la Biodiversidad, Instituto de Biología, Universidad Nacional Autónoma de México.
Molecular analysis
Two bunocotylids were placed individually overnight in tubes with a digestion solution for DNA extraction at 56°C. The digestion solution contained 10 mM Tris-HCl (pH 7.6), 20 mM NaCl, 100 mM Na2 EDTA (pH 8.0), 1 % sarkosyl, and 0.1 mg/ml proteinase K. DNA was extracted using DNAzol reagent (Molecular Research Center, Cincinnati, Ohio). The domains D1–D3 of the large subunit of nuclear ribosomal RNA gene (28S) were amplified via PCR using the primers: 391 5′– AGCGGAGGAAAAGAAACTAA–3′, and 536: 5′-CAGCTATCCTGAGG GAAAC-3′ (García-Varela and Nadler, 2005). The amplification and sequencing protocols followed those used in Andrade-Gómez et al. (2023). Sequences were assembled and edited using Geneious v7 (Kearse et al., 2012) and deposited in the GenBank database.
The two newly obtained sequences were aligned with data from other members of Hemiuroidea downloaded from the GenBank dataset, plus one species from Azygiidae used as outgroup for rooting the tree (see Table 1). The final alignment consisted of 35 sequences with 1,326 nucleotides. Alignments were built using the software Clustal W (Thompson et al., 1997) with default parameters and adjusted manually with the Mesquite software (Maddison & Maddison, 2011).
Phylogenetic analyses were performed using Maximum Likelihood (ML) and Bayesian Inference (BI) methods. ML was carried out with RAxML version 7.0.4 (Silvestro & Michalak, 2011) and Bayesian Inference analyses were run with MrBayes version 3.2.7 (Ronquist et al., 2012) using the online interface CIPRES (Cyberinfrastructure for Phylogenetic Research) Science Gateway v3.3 (Miller et al., 2010). The best model was estimated with the Akaike information criterion (AIC) using the jModel Test version 0.1.1 program (Posada, 2008), which predicted the best model for the 28S dataset to be GTR + I + G. Nodal ML support was achieved through 1000 bootstrap replicates. The BI analyses included Markov Chain Monte Carlo (MCMC) searches of two simultaneous runs for 10 million generations, with sampling every 1000 generations, a heating parameter value of 0.2, and the first 25% of the sampled trees were discarded. Trees were drawn using FigTree program v.1.4.4 (Rambaut, 2010). Uncorrected P distances were obtained in MEGA version 6 (Tamura et al., 2011).
Results
Class Trematoda Rudolphi, 1808
Subclass Digenea Carus, 1863
Order Plagiorchiida La Rue, 1957
Suborder Hemiurata Skrjabin & Guschanskaja, 1954
Superfamily Hemiuoroidea Looss, 1899
Family Bunocotylidae Dollfus, 1950
Parasaturnius n. gen.
Type-species: Saturnius maurepasi Overstreet, 1977.
Diagnosis. Body elongate, cylindrical, with maximum width at ventral sucker flange level. Tegument unarmed, with fine transverse striations. Two circular muscular flanges present around body. Anteriormost flange surrounds oral sucker. Second flange at posterior end of ventral sucker, forms 2 lateral subconical protuberances with concentric muscles. Body with 7 pseudosegments separated by 6 transverse fibrous septa. Oral sucker muscular, subterminal bearing small papillae. Large single cells present in segments.
Ventral sucker strongly muscular, sub-spherical, anterior to mid-body. Prepharynx absent; pharynx subspherical. Oesophagus short. ‘Drüsenmagen’ present. Caeca with constrictions at septa levels. Testes 2, smooth, in tandem, located at hindbody. Seminal vesicle large, saccular antero-dorsal to ventral sucker, larger than sinus-sac. Sinus-sac small elongate-oval, contains short muscular hermaphroditic duct. Pars prostatica vesicular, small, difficult to distinguish, enters sinus-sac at its base. Genital pore at level of anterior septum. Ovary transverse, oval, in anterior end of last pseudosegment, ventral to caeca, smooth to irregular, contiguous with vitellarium. Mehlis’ gland and uterine seminal receptacle present. Laurer’s canal not observed. Vitellarium compact, smooth to irregular, elongate-oval, larger than ovary. Uterus thin-walled, extends posterior to vitellarium. Metraterm short. Eggs numerous. Excretory pore wide, terminal or subterminal; vesicle Y-shaped.
Etymology. The genus Parasaturnius n. gen. refers to its resemblance with Saturnius Manter, 1969, and uses the Greek prefix para (meaning resemble).
Remarks
Parasaturnius n. gen. can be differentiated from Robinia and Bunocotyle by the presence of pseudosegments (transverse fibrous septa) in the fore- and hind-body. Furthermore, the genus Robinia possesses fine striations, symmetrical testes, 11-15 muscular lobes on the oral sucker, and vestigial ecsome. The new genus differs further from Bunocotyle by the lack of cyclocoel. Parasaturnius n. gen. is morphologically very similar to Saturnius since both genera present pseudosegments. Nonetheless, the new genus can be distinguished by the presence of four pairs of small papillae surrounding the aperture of the oral sucker.
Parasaturnius maurepasi (Overstreet, 1977) n. gen. n. comb. (Fig. 1)
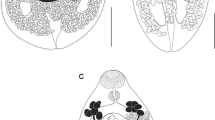
Parasaturnius maurepasi (Overstreet, 1977) n. gen. n. comb. from Mugil curema (A) whole worm voucher (Dorsal view); Scanning electron micrographs of voucher (B) Whole worm; (C) Forebody (D) Oral sucker, Arrows indicates papillae; (E) Ventral sucker. Scale bars (μm) = (A, B) 100; (C) 50; (D,E) 10.
Syn. Saturnius maurepasi Overstreet, 1977
Records: 1. Overstreet (1977); 2. Romero & Galeano (1981); 3. Fernandes & Goulart (1992); 4. Knoff et al. (1997); 5. Present study.
Type host: Mugil cephalus L. (1, 2)
Other hosts: Mugil liza Valenciennes, parati mullet (3, 4); Mugil curema Valenciennes, silver mullet (5)
Type locality: Ocean Springs, Mississippi (1)
Other localities: off Santa Marta, Colombia (2); Brazilian coast of SW Atlantic (3, 4); La Carbonera, Yucatán State, Mexico (5)
Site of infection: Stomach lining
Prevalence: 70% (5)
Intensity: 1-6 (5)
Specimens studied: 10 voucher specimens (CNHE 12843)
Representative DNA sequences: OR831227– OR831228 (28S)
Redescription (Based on 10 mature individuals. Measurements in Table 2)
Body elongate, gradually tapering anteriorly, with seven pseudosegments. Maximum body width at level of ventral sucker flange, width 9.2−16.8 of body length. Preoral dorsal lip wide (Fig 1d). Two flanges, one located at oral sucker level, and the second at or slightly posterior to ventral sucker, mound-shaped. Oral sucker aperture surrounded by 8 small papillae arranged in four pairs (Fig 1d). Ventral sucker located between the second and third pseudosegment. Forebody 25.4−35.8% of body length. Large single cells in most segments, lateral to body. Pharynx contiguous; prepharynx not observed. Oesophagus thick walled, smaller than pharynx. Caeca with constrictions in pseudosegments, terminating near posterior end, “Drüsenmagen” just posterior to oesophagus.
Testes 2, smooth or irregular, in the last two segments, in tandem, between the caeca. Sinus sac small containing eversible hermaphroditic duct; metraterm short. Seminal vesicle large, elongate, longer than sinus sac, extending to anterior border of ventral sucker. Pars prostatica vesicular, small, difficult to observe, enters sinus-sac as its base. Genital pore median at or near level of anterior septum.
Ovary transversely elongate, pos- testicular, in last major segment, ventral, smooth to irregular, in middle of caeca. Mehlis’ gland conspicuous in some specimens. Vitellarium smooth, 7.9−12.6 of body length, contiguous with ovary. Uterus filling most of partitions, extending posterior to vitellarium but not reaching posterior end. Eggs numerous, small. Excretory vesicle consisting of posterior muscular sac within the last pseudosegment. Pore terminal.
Remarks
The specimens found in Mugil curema from La Carbonera coastal lagoon were identified as Parasaturnius maurepasi n. gen. n. comb. based on morphology, host and geographical distribution as in the original description (Overstreet, 1977) (Table 2). In addition, following the key to species proposed by Blasco-Costa et al. (2008), our specimens fall in S. maurepasi since they possess seven pseudosegments separated by six transverse fibrous septa. However, we found some slight morphological differences with respect to the original description, as the papillae surrounding the oral sucker, since we were able to obtain SEM photomicrographs of the sampled specimens. Moreover, the metrical data suggest specimens found in the present study in Yucatán are overall slightly smaller than the specimens described by Overstreet (1977).
Molecular data and phylogenetic analysis
The 28S data set included 35 sequences and comprised 1, 326 nucleotides. The alignment (trimmed to the shortest sequence) included one sequence of Azygiidae used as outgroup. The phylogenetic analyses inferred with ML and BI recovered similar topologies (Fig. 2). The analyses show that the superfamily Hemiuroidea is monophyletic. Within each family, Hemiuridae, Isoparorchiidae, and Derogenidae were recovered as monophyletic, with strong nodal support (1/100; 1/97; 0.99/94). The remaining three families analyzed were not recovered as monophyletic, e. g., Gonocercidae, Lecithasteridae and Bunocotylidae. Bunocotylids were separated in two independent clades, one formed by three subfamilies, Hysterolecithinae, Quadrifoliovariinae and Opisthadeninae, with low support (0.54/48). The second major clade was formed only by Bunocotylinae, with strong support (1/100). Within Bunocotylinae, the two newly generated sequences of Parasaturnius maurepasi n. gen. n. comb. were nested as the sister group of the three genera contained in the subfamily, e.g., Robinia, Bunocotyle, and Saturnius, with strong support (1/100) (Fig. 2).
Consensus Bayesian Inference and Maximum Likelihood trees of genera of Hemiuroids inferred with 28S rRNA sequence data; numbers near internal nodes show posterior probabilities (BI) and ML bootstrap clade frequencies. Opis= Opisthadeninae; Quad=Quadrifoliovariinae; Hyst= Hysterolecithinae. HEM= Hemiuridae, ISO=Isoparorchiidae; DERO= Derogenidae; GON= Gonocercidae; LEC= Lecithasteridae; FU=Family Unknown sensu Atopkin et al. 2020.
The genetic divergence estimated for 28S for the Bunocotylinae and the other three subfamilies (Hysterolecithinae, Opisthadeninae, Quadrifoliovariinae) ranged from 12.6 to 19.1%. Within Bunocotylinae, the two new isolates of Parasaturnius maurepasi n. gen. n. comb. varied 10.7-12% with Saturnius spp., 10.5-11.5% with B. progenetica, and 10.8-11.5 with R. aurata. The intraspecific genetic divergence between the two isolates of the new species we described in this study was 0.2% (Table 3).
Discussion
The two newly sequenced specimens of Parasaturnius maurepasi n. gen. n. comb. were recovered as the sister group of the three previously mentioned genera, with the same topology from previous studies, e.g., Robinia as sister of Saturnius and Bunocotyle (Pankov et al., 2006; Marzoug et al., 2014). Interestingly, even though Saturnius and Parasaturnius n. gen. are morphologically very similar, molecularly they are not closely related and exhibit large genetic divergence of the gene 28S. Saturnius gibsoni (KJ010542) and S. minutus (DQ354366) are sister taxa to Bunocotyle progenetica (DQ354365), with less than 3% of genetic divergence, which is quite low if we consider that divergence between Robinia aurata and the three species previously mentioned varies from 6.6 to 6.8% (Table 3). Furthermore, the genetic divergence between Parasaturnius maurepasi n. gen. n. comb. and the four species was even higher, and ranged from 10.7–12%, which suggest that presence of pseudosegments in Saturnius and the new genus are homoplasies in these two genera. Interestingly, these high genetic divergence values between genera are like those reported among other genera of hemiuroid trematodes (Pantoja & Kudlai, 2022).
The original description of P. maurepasi n. gen. n. comb. was published by Overstreet (1977) (as Saturnius maurepasi) from the stomach of Mugil cephalus in Ocean Springs, Mississippi, USA. After the original record, the species has been reported across the Atlantic coast of the Americas, as a parasite of M. liza in Colombia and Brazil (Romero & Galeano, 1981; Fernandes & Goulart, 1992; Knoff et al., 1997). The species is reported for the first time in Mexico, as a parasite of Mugil curema in La Carbonera coastal lagoon, representing new host and new locality records for this parasite. In the same geographic region, a morphologically similar species has been reported, i.e., S. belizensis. Fischtal (1977) described S. belizensis as a parasite of M. curema from Belize, a locality relatively close to La Carbonera, Yucatán, Mexico. It is noteworthy that most probably both, Overstreet (1977) and Fischtal (1977), did not know about each other´s paper describing a new species of Saturnius published in the same year. Furthermore, S. belizensis was described from only two specimens. In the last taxonomic review of the genus Saturnius, Blasco-Costa et al. (2008) studied a paratype of this species and noticed that S. belizensis exhibits five pseudogements. Even though our specimens are metrically more similar to S. belizensis, the number of pseudosegments in our specimens are seven, similar to that reported by Blasco-Costa et al. (2008) for S. maurepasi. Testing the validity of S. belizensis will require the generation of 28S DNA sequences from the type locality, and a through morphological study of a larger number of specimens to corroborate the number of pseudosegments. In addition to that, the schematic drawings of S. maurepasi from M. liza in Brazil shows they possess six pseudosegments; based on host association and geographical distribution, these specimens could indeed represent a separate lineage. Therefore, a detailed morphological study and sequence data are required to corroborate the status of these reports (Fernandes & Goulart, 1992).
Based on the evidence discussed above, Parasaturnius maurepasi n. gen. n. comb. is here recognized based on molecular data and a detailed morphological analysis. Still, sampling specimens of other valid species is required to be analyzed under a molecular approach, to clarify the species and family status. It is necessary to study further Saturnius segmentatus, the type species, which is the only species allocated in the genus Saturnius exhibiting morphological traits similar to those of Robinia, such as the muscular and conspicuous papillae on the oral sucker. In addition, the cyclocoel is a morphological trait of Bunocotyle, however this character is also found in four species of Saturnius, i.e., S. gibsoni, S. minutus, S. dimitrovi and S. overstreeti. In this sense, it would be also necessary to obtain sequence data of Saturnius spp. lacking cyclocoel and Bunocotyle spp. to test the evolution of morphological traits in Bunocotylinae.
Comments on the taxonomic status of Bunocotylidae.
Using molecular data, Sokolov et al. (2018) and Atopkin et al. (2020) resurrected the family Bunocotylidae, and considered it contained the subfamilies Bunocotylinae Opisthadeninae, Hysterolecithinae and Quadrifoliovariinae. However, two recent studies provided evidence in favor of the paraphyly of Bunocotylidae (Louvard et al., 2022; Faltýnkova et al., 2022), although these studies did not include all the genera considered by Atopkin et al. (2020) as members of the family. In our study, after including all the genera of bunocotylids following the concept by Atopkin et al. (2020) we found no conclusive evidence on the interrelationships among members of the families Bunocotylidae, Lecithasteridae and Hemiuridae. The phylogenetic tree yielded a basal polytomy and it might be premature to consider that the genera Machidatrema León-Règagnon, 1998, Hysterolecithoides Yamaguti, 1934, Bilacinia Manter, 1969, Quadrifoliovarium Yamaguti, 1965, Opisthadena Linton 1910, and Genolinea Manter, 1925 (allocated in Hysterolecithinae, Opisthadeninae, Quadrifoliovariinae) belong to Bunocotylidae. Therefore, more sequence data is required to resolve the systematic interrelationships among this hemiuroid group.
References
Andrade-Gómez, L., Ortega-Olivares, M.P., Solórzano-García, B., García-Varela, M., Mendoza-Garfias, B., & Pérez-Ponce de León, G. (2023). Monorchiids (Digenea, Trematoda) of fishes in the Yucatán Peninsula, Mexico, with the description of three new species based on morphological and molecular data. Parasite, 30, 15. https://doi.org/10.1051/parasite/2023015
Atopkin, D. M., Besprozvannykh, V. V., Beloded, A. Yu., Ngo, H. D., Ha, N. V., & Tang, N. V. (2017). Phylogenetic relationships of Hemiuridae (Digenea: Hemiuroidea) with new morphometric and molecular data of Aphanurus mugilis Tang, 1981 (Aphanurinae) from mullet fish of Vietnam. Parasitology International, 66, 824–830. https://doi.org/10.1016/j.parint.2017.09.009
Atopkin, D. M., Nakao, M., Besprozvannykh, V.V., Ha, N.D., Nguyen, H.V., & Sasaki M. (2020) Morphological and molecular data for species of Lecithaster Lühe, 1901 and Hysterolecithoides Yamaguti, 1934 (Digenea: Lecithasteridae) from fish of East Asia and phylogenetic relationships within the Hemiuroidea Looss, 1899. Journal of Helminthology, 94, 1–13. https://doi.org/10.1017/S0022149X18001049.
AVMA (American Veterinary Medical Association). 2020. Guidelines for the euthanasia of animals. Schaumburg, Illinois: American Veterinary Medical Association.
Besprozvannykh, V. V., Atopkin, D. M., Ngo, H. D., Ermolenko, A. V., Van, Ha. N., Van Tang, N., & Beloded, A. Yu. (2017). Morphometric and molecular analyses of two digenean species in mugilid fish: Lecithaster mugilis Yamaguti, 1970 from Vietnam and L. sudzuhensis n. sp. from southern Russian Far East. Journal of Helminthology, 91, 326–331. https://doi.org/10.1017/S0022149X16000201
Blasco-Costa, I., Pankov, P., Gibson, D. I., Balbuena, J. A., Raga, J. A., Sarabeev, V. L., & Kostadinova, A. (2006). Saturnius minutus n. sp. and S. dimitrovi n. sp. (Digenea: Hemiuridae) from Mugil cephalus L. (Teleostei: Mugilidae) with a multivariate morphological analysis of the Mediterranean species of Saturnius Manter, 1969. Systematic Parasitology, 65, 77–91. https://doi.org/10.1007/s11230-006-9043-9
Blasco-Costa, I., Montero, F. E., Gibson, D. I., Balbuena, J. A., Raga, J. A., Shvetsova, L. S., & Kostadinova, A. (2008). A revision of the species of Saturnius Manter, 1969 (Digenea: Hemiuridae), parasites of mullets (Teleostei: Mugilidae). Systematic Parasitology, 71, 53–74. https://doi.org/10.1007/s11230-008-9141-y
Calhoun, D. M., Curran, S. S., Pulis, E. E., Provaznik, J. M., & Franks, J. S. (2013). Hirudinella ventricosa (Pallas, 1774) Baird, 1853 represents a species complex based on ribosomal DNA. Systematic Parasitology, 86, 197–208. https://doi.org/10.1007/s11230-013-9439-2
Chambers, C. B., & Cribb, T. H. (2006). Phylogeny, evolution and biogeography of the Quadrifoliovariinae Yamaguti, 1965 (Digenea: Lecithasteridae). Systematic Parasitology, 63, 61–82. https://doi.org/10.1007/s11230-005-9007-5
Claxton, A. T., Fuehring, A. D., Andres, M. J., Moncrief, T. D., & Curran, S. S. (2017). Parasites of the vermilion snapper, Rhomboplites aurorubens (Cuvier), from the Western Atlantic Ocean. Comparative Parasitology, 84, 1–14. https://doi.org/10.1654/1525-2647-84.1.1
Faltýnkova, A., Kudlai, O., Salganskiy, O.O., Korol, E.M., & Kuzmina T.A. (2022) Trematodes from Antarctic teleost fishes off Argentine islands, West Antarctica: molecular and morphological data. Systematic Parasitology, 99, 491–523, https://doi.org/10.1007/s11230-022-10041-9.
Fernandes, B. M. M., & Goulart, M. B. (1992). First report of the genera Macvicaria Gibson & Bray, 1982, Pachycreadium Manter, 1954 and Saturnius Manter, 1969 (Trematoda: Digenea), in Brazilian marine fishes. Memórias do Instituto Oswaldo Cruz, 87 (Supplement 1), 101–104.
Fischtal, J. H. (1977). Some digenetic trematodes of marine fishes from the barrier reef and reef lagoon of Belize. Zoologica Scripta, 6, 81–88.
García-Varela, M., & Nadler, S. A. (2005). Phylogenetic relationships of Palaeacanthocephala (Acanthocephala) inferred from SSU and LSU rRNA gene sequences. Journal of Parasitology, 91, 1401–1409. https://doi.org/10.1645/GE-523R.1
Kearse, M., Moir, R., Wilson, A., Stones-Havas, S., Cheung, M., Sturrock, S., Buxton, S., Cooper, A., Markowitz, S., Duran, C., Thierer, T., Ashton, B., Meintjes, P., & Drummond, A. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Knoff, M., Luque, J. L., & Amato, J. F. R. (1997). Community ecology of the metazoan parasites of grey mullets, Mugil platanus (Osteichthyes: Mugilidae) from the littoral of the state of Rio de Janeiro, Brazil. Revista Brasileira de Biologia, 57, 441–454.
Kohn, A, Fernandes, B. M., & Cohen, S. C. (2007). South American trematodes parasites of fishes. Fiocruz, Instituto Oswaldo Cruz/Imprinta Express Ltd: Rio de Janeiro.
Louvard, C., Cutmore, S.C., Yong, R.Q.Y., Dang, C.& Cribb, T.H. (2022) First elucidation of a didymozoid life cycle: Saccularina magnacetabula n. gen. n. sp. infecting an arcid bivalve, International Journal of Parasitology, 52, 407–425. https://doi.org/10.1016/j.ijpara.2021.11.012.
Maddison, W.P., & Maddison, D. R. (2011). Mesquite: a modular system for evolutionary analysis. Version 2.75. Available at http://mesquiteproject.org
Martin, S. B., De Silva, M. L. I, Pathirana, E., & Rajapakse, R. P. V. J. (2023) Polyphyly of the Dinurinae Looss, 1907 (Digenea: Hemiuridae) and resurrection of the Mecoderinae Skrjabin & Guschanskaja, 1954 based on novel collection of Tubulovesicula laticaudi Parukhin, 1969 from marine elapid snakes in Sri Lanka. Parasitology International, 97, 102776. https://doi.org/10.1016/j.parint.2023.102776
Marzoug, D., Rima, M., Boutiba, Z., Georgieva, S., Kostadinova, A., & Pérez-del-Olmo, A. (2014). A new species of Saturnius Manter, 1969 (Digenea: Hemiuridae) from Mediterranean mullet (Teleostei: Mugilidae). Systematic Parasitology, 87, 127–134. https://doi.org/10.1007/s11230-013-9468-x
Miller, A. M., Pfeiffer, W., & Schwartz, T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. Gateway Computing Environments Workshop (GCE). p. 1–8.
Olson, P. D., Cribb, T. H., Tkach, V. V., Bray, R. A., & Littlewood, D. T. J. (2003). Phylogeny and classification of the Digenea (Platyhelminthes: Trematoda). International Journal for Parasitology, 33, 733–755. https://doi.org/10.1016/S0020-7519(03)00049-3
Overstreet, R. M. (1977). A revision of Saturnius Manter, 1969 (Hemiuridae: Bunocotylinae) with descriptions of two new species from the striped mullet. Universidad Nacional Autónoma de México, Instituto de Biología Publicaciones Especiales, 4, 273–284.
Pankov, P., Webster, B. L., Blasco-Costa, I., Gibson, D. I., Littlewood, D. T. J., Balbuena, J. A., & Kostadinova, A. (2006). Robinia aurata n. g., n. sp. (Digenea: Hemiuridae) from the mugilid Liza aurata with a molecular confirmation of its position within the Hemiuroidea. Parasitology, 133, 217–227. https://doi.org/10.1017/S0031182006000126
Pantoja, C., & Kudlai, O. (2022). Hemiurid Trematodes (Digenea: Hemiuridae) from Marine Fishes off the Coast of Rio de Janeiro, Brazil, with Novel Molecular Data. Animals,12, 3355. https://doi.org/10.3390/ani12233355
Posada, D. (2008). jModelTest: phylogenetic model averaging. Molecular Biology and Evolution, 25, 1253–1256. https://doi.org/10.1093/molbev/msn083
Rambaut, A. (2010) FigTree v1.4.4. Institute of Evolutionary Biology, University of Edinburgh, Edinburgh.http://tree.bio.ed.ac.uk/software/figtree/
Romero, M., & Galeano, M. L. (1981). Contribución al conocimiento de parásitos de peces de la ciénaga de Santa Marta (Familias Hemiuridae y Allocreadiidae: Trematoda Digenea). Lozanía, 34, 5–8.
Ronquist, F., Teslenko, M., van der Mark, P., Ayres, D. L., Darling, A., Hohna, S., Larget, B., Liu, L., Suchard, M. A., & Huelsenbeck, J. P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. https://doi.org/10.1093/sysbio/sys029
Silvestro, D., & Michalak, I. (2011). RaxmlGUI: a graphical front-end for RAxML. Organisms Diversity & Evolution, 12, 335–337. https://doi.org/10.1007/s13127-011-0056-0
Sokolov, S. G., Atopkin, D. M., Gordeev, I. I., & Shedko, M. B. (2018) Phylogenetic position of the genus Gonocerca Manter, 1925 (Trematoda, Hemiuroidea), based on partial sequences of 28S rRNA gene and a reconsideration of taxonomic status of Gonocercinae Skrjabin et Guschanskaja, 1955. Parasitology International 67, 74–78. https://doi.org/10.1016/j.parint.2017.03.007
Sokolov, S. G., Atopkin, D. M., Urabe, M., & Gordeev, I. I. (2019). Phylogenetic analysis of the superfamily Hemiuroidea (Platyhelminthes, Neodermata: Trematoda) based on partial 28S rDNA sequences. Parasitology, 146, 596–603. https://doi.org/10.1017/S0031182018001841
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., & Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution, 28, 731–739. https://doi.org/10.1093/molbev/msr121
Thompson, J. D., Gibson, T. J., Plewniak, F., & Jeanmougin, F. (1997). The Clustal windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research, 25, 4876–4882. https://doi.org/10.1093/nar/25.24.4876
WoRMS (2023). Bunocotylidae Dollfus, 1950. Accessed at: https://www.marinespecies.org/aphia.php?p=taxdetails&id=344841 on 2023-08-27cc
Acknowledgments
LAG thanks to the Dirección General de Asuntos de Personal Académico (DGAPA-UNAM) Mexico for the Postdoctoral Fellowship granted. We are truly grateful to Berenit Mendoza Garfias, LaNaBio for the help obtaining the SEM images. We are grateful to Laura Marquez and Nelly López, LaNaBio for their help for sequencing DNA. We thank Luis García Prieto for providing access numbers of CNHE. We thank to Norberto Colín for the use of the facility of the Molecular Biology lab at ENES-Mérida. We sincerely thank Maribel Badillo Alemán and Alfredo Gallardo Torres, Laboratorio de Biología de la Conservación, Facultad de Ciencias UNAM for lab facilities and the identification of the host.
Funding
This research was supported by grants from the Programa de Apoyo a Proyectos de Investigación e Inovación Tecnológica (PAPIIT-UNAM IN212621), and the Consejo Nacional de Ciencia y Tecnología (CONACYT A1-S-21694) to GPPL.
Author information
Authors and Affiliations
Contributions
LAG and GPPL wrote the main text LAG prepared the figures
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical standards
Specimens were collected under the sampling permit granted to the Laboratorio de Biología de la Conservación (BioCon) by the Comisión Nacional de Acuacultura y Pesca (CONAPESCA), No. PPF/DGOPA-001/20. Fish were humanely euthanized following the protocols described by the 2020 edition of the AVMA Guidelines for the Euthanasia of Animals.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Andrade-Gómez, L., Pérez-Ponce de León, G. Parasaturnius maurepasi n. gen. et n. comb. (Digena: Bunocotylidae) from the stomach of the silver mullet, Mugil curema (Perciformes: Mugilidae) in coastal lagoons of northern Yucatán, Mexico. Syst Parasitol 101, 16 (2024). https://doi.org/10.1007/s11230-023-10142-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11230-023-10142-z