Abstract
At present, many cloning selection algorithms have been studied, and improvements have been made to the cloning, mutation and selection steps. However, there is a lack of research on the optimization of the updating operation steps. The clonal selection algorithm is traditionally updated through a random complement of antibodies, which is a blind and uncertain process. The added antibodies may gather near a local optimal solution, resulting in the need for more iterations to obtain the global optimal solution. To solve this problem, our improved algorithm introduces a crowding degree factor in the antibody updating stage to determine whether there is crowding between antibodies. By eliminating antibodies with high crowding potential and poor affinity, the improved algorithm guides the antibodies to update in the direction of the global optimal solution and ensures stable convergence with fewer iterations. Experimental results show that the overall performance of the improved algorithm is 1% higher than that of the clonal selection algorithm and 2.2% higher than that of the genetic algorithm, indicating that the improved algorithm is effective. The improved algorithm is also transplanted to other improved clonal selection algorithms, and the overall performance is improved by 0.97%, indicating that the improved algorithm can be a beneficial supplement to other improved clonal selection algorithms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The clonal selection algorithm (CSA) [1] was proposed based on the eponymous principle of biological systems that explains the immune response of B cells in organisms during the recognition of nonself antigens [2]. The clonal selection algorithm is a heuristic optimization algorithm that involves cloning and mutation of a single antibody to expand the search range. As a heuristic algorithm to solve complex problems with a high risk of mutation, the clonal selection algorithm converges quickly and has obvious advantages over other current mainstream intelligent optimization algorithms [3]. In recent years, it has been applied to scheduling problems [4, 5], fault diagnosis [6], function optimization [7], parameter recognition [8], etc.
However, when solving practical problems, the traditional clonal selection algorithm has problems with algorithm efficiency and convergence speed, and it easily falls into local optima [9]. The process of the clonal selection algorithm mainly includes cloning, mutation, selection, updating and other steps. At present, many scholars’ improvements of clonal selection algorithms mainly focus on the optimization of key steps such as cloning, mutation and selection to improve the convergence efficiency of the algorithm. For cloning steps, Song Dan et al. [10] designed a search mechanism based on fuzzy nongenetic information, combined it with the clonal selection principle, and proposed a double clonal selection algorithm based on fuzzy nongenetic information memory, which effectively reduced the number of virtual collisions in evolutionary iteration and improved the global convergence speed and precision. Lingjie Li et al. [11] proposed a multiobjective immune algorithm cloning mechanism based on vertical distance, which was used to dynamically arrange the number of clones of each antibody to improve the population diversity. Akemi Galveza et al. [12] proposed an adaptive elite clonal selection algorithm for a multimodal and multivariate continuous nonlinear optimization problem. After the cloning steps of the algorithm were improved, cloning was no longer dependent on the affinity ratio, and all antibodies were cloned with the same probability, thus solving the optimization problem successfully. In terms of variation steps, reference [13] generates variation factors through a cloud model, improves variant antibodies, and selects clonal variant antibodies according to a reverse learning strategy to improve the global optimization ability and search efficiency of the algorithm. Yi Wang et al. [14] proposed an adaptive clonal selection algorithm with multiple differential change strategies using their historically documented pool of adaptive mutation strategies to guide the immune response process and speed up convergence through an adaptive population size adjustment method. For the mutation and selection steps, Shu Wanneng et al. [15] integrated the simulated annealing algorithm into the clonal selection algorithm, carried out mutation and selection operations according to the annealing probability, effectively maintained a balance between global search and local search, improved the search efficiency of the algorithm and accelerated the convergence rate of the algorithm.
As seen from the above literature, a large number of algorithms have been studied and used to improve cloning, mutation, selection, etc., but there is a lack of research on the optimization of updating operation steps. The clonal selection algorithm sorts according to affinity, clones and mutates the first N antibodies with high affinity, and randomly updates the remaining d antibodies. Random updating causes problems such as blindness and uncertainty. If the position of the added antibody is close to the global optimal solution or the affinity of the added antibody is high, there is a high probability that the global optimal solution can be obtained quickly. If the added antibody is close to a local optimal solution or the antibodies gather near a local optimal solution, the probability of obtaining the global optimal solution decreases, and more iterations may be needed to obtain the global optimal solution. The position and affinity of the added antibody are random, resulting in greater randomness in the final result, and the algorithm performance is not stable. Therefore, we believe that the blind random update restricts the algorithm performance, which easily leads to slow algorithm convergence, local optimization, poor algorithm stability and other problems. To compensate for the lack of research regarding this topic, inspired by the idea of using affinity distance correlation to analyze the characteristics of optimization problems in the literature [16], we propose a new strategy to update the antibody purposefully and effectively so that the algorithm has faster convergence speed, higher optimization accuracy and better convergence stability.
This paper presents an improved clonal selection algorithm based on the directed update strategy clonal selection algorithm (DUSCSA), which focuses on improving the update operation of the clonal selection algorithm based on previous studies. In this paper, we refer to the phenomenon of antibody clustering caused by random updates of the algorithm as “crowding.” During “crowding,” the antibodies are clustered around the local optimal solution with a high probability of converging to the local optimal solution, while convergence to the global optimal solution requires more iterations. The basic idea of the algorithm is to set a minimum distance threshold to determine whether the complementary antibodies are “crowded” and to eliminate antibodies that are “crowded” and have poor affinity with the optimal antibodies in the current iteration so that the complementary antibodies are updated along a direction that is closer to the global optimal solution, increasing the chance of obtaining the global optimal solution and ensuring a stable convergence of the algorithm. The method in this paper provides a new perspective on the update phase of the clonal selection algorithm and is a useful addition to other clonal selection improvement algorithms. Our results can be transferred to improve the convergence and efficiency of other algorithms.
2 Overview of the clonal selection algorithm
2.1 Principle of the clonal selection algorithm
Organisms have a variety of immunologically active cells, and immune cells undergo mitosis to produce clones, i.e., a single cell gives rise to a group of cells. This mechanism of generating antibodies that recognize and destroy invading antigens and using asexually reproducing clones to activate differentiated and proliferating antibodies to achieve an immune response and ultimately remove the antigen is called clonal selection [17]. Based on this mechanism, clonal selection optimization algorithms have evolved to solve function optimization problems. The correspondence between clonal selection and problem solving in biological systems is shown in Table 1.
2.2 Clonal selection algorithm flow
The flow of the traditional CSA is shown in the following steps:
-
Step 1: Initialize the parameter. Initialize the population size D, outstanding antibody size N, dimension dim, upper and lower bounds M and m for the array of antibody values, respectively, maximum number of iterations \(G_{max}\), and variation probability p where \(N < D\).
-
Step 2: Initialize the antibody population. Random generated antibody population of antibodies A of size D, \(x_i\) is the i th antibody x, \(i \in [1, D]\).
$$\begin{aligned} A&= \{x_1,x_2,\ldots ,x_i,\ldots ,x_D\} \end{aligned}$$(1) -
Step 3: Calculate affinity. Calculate the affinity of each antibody in the antibody population by using the objective function as the target value.
-
Step 4: Perform segmentation. Sort the calculated antibody affinities and select the top N antibodies as the outstanding antibody population B.
$$\begin{aligned} B&= \{x_1,x_2,\ldots ,x_i,\ldots ,x_N\} \end{aligned}$$(2) -
Step 5: Clone. Clone each antibody at different scales, depending on affinity, to obtain the clonal population \(B'\).
$$\begin{aligned} B'&= \{x_1,\ldots ,x_1,x_2,\ldots ,x_2,x_i,\ldots ,x_i,\ldots ,x_N\ldots ,x_N\} \end{aligned}$$(3) -
Step 6: Mutate. Mutate the \(B'\) antibody population with probability p to obtain the mutated antibody population \(B''\), where \(x'_i\) is the i th mutated antibody x, \(i \in [1,N]\).
$$\begin{aligned} B''&= \{x'_1,\ldots ,x'_1,x'_2,\ldots ,x'_2,x'_i,\ldots ,x'_i,\ldots ,x'_N\ldots ,x'_N\} \end{aligned}$$(4) -
Step 7: Select. For the cloned mutated antibodies, the antibody with the best affinity is selected to replace the original antibody. Finally, the new top N antibodies with good affinity are obtained as a new population of outstanding antibodies C.
$$\begin{aligned} C&= \{x'_1,x'_2,\ldots ,x'_i,\ldots ,x'_N\} \end{aligned}$$(5) -
Step 8: Stock update. Randomly replenish d antibodies combined with the outstanding antibody population C as the initial population for the next generation.
$$\begin{aligned} d&= D-N \end{aligned}$$(6) -
Step 9: Repeat step 3 if the maximum number of iterations \(G_{max}\) is not satisfied; otherwise, output the result.
2.3 Thoughts on the clonal selection algorithm
As seen from the flow of the clonal selection algorithm, after the first N good antibodies are divided in step 4, steps 5, 6 and 7 are all about cloning, mutation and selection of these N antibodies, respectively. Step 8 replaces the last d antibodies delineated in step 4 by randomly complementing them. For ease of description, as shown in Fig. 1, cross sections of a three-dimensional image of a complex function, as shown in Fig. 2, which possesses a global optimal solution and a separate local optimal solution, are taken and described by dividing them into three regions according to their affinity to the global optimal solution. The first tier comprises the antibodies with the closest affinities to the global optimal solution; the second tier comprises the antibodies with worse affinities than the first tier and better affinities than the third tier; and the third tier comprises the antibodies with the worst affinity.
With random updating, there are two possible scenarios for the distribution of the positions of the complemented antibodies, as shown in Fig. 2: first, the complemented antibodies are near the local optimal solution, and after selecting the first N antibodies and eliminating the last d antibodies, as shown in Fig. 3, the antibodies within the second layer are retained. The antibodies are all clustered near the local optimal solution, which is not conducive to obtaining the global optimal solution. Second, the antibodies that are added are near the global optimal solution, and if these antibodies are selected to participate in the next round of iterations for clonal variation, the algorithm has an increased chance of obtaining the global optimal solution. As shown in Fig. 4, the complemented antibodies have poor affinities and do not enter the second region, and most of the antibodies in the antibody pool remain clustered around the local optimal solution. Both cases result in the antibodies clustering around the local optimal solution with a high probability of converging to the local optimal solution, and convergence to the global optimal solution thus requires a higher number of iterations. Random updating is blind and uncertain; if the positions of the complementary antibodies are close to the global optimum or if the affinities of the complementary antibodies are high, there is a high probability of rapid convergence to the global optimum. Therefore, blind random updating makes the convergence of the algorithm unstable.
Based on the above analysis of the problems associated with random updating, if the randomly added antibodies are positioned close to the global optimal solution or if the added antibodies have a high affinity, the probability of obtaining the global optimal solution is higher and convergence is faster. By directing the randomly added antibodies to be updated in a direction close to the global optimum, the algorithm converges steadily and quickly, and the accuracy of the search is higher.
3 Improved clonal selection algorithm
3.1 Update operations with a targeted update policy
The directed update strategy is an improvement on the traditional random update method. The integration of the directed update strategy into the update operation of the clonal selection algorithm regulates the balance between randomness and determinism, thus avoiding the problems of low accuracy in finding the best solution and slow convergence rate due to complete randomness in the local search phase [18]. It can also avoid the blindness and uncertainty of antibodies generated by complete randomness, which may gather near a local optimal solution and produce crowding, so that the global optimum is never found.
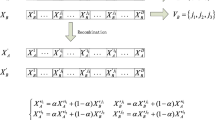
The DUSCSA uses a threshold \(\sigma \) to determine whether to add a random complement of antibodies to the antibody population. The threshold calculates the farthest distance between antibodies and introduces a crowding degree factor (cdf), where cdf \(\in (0,1),\) which is used as the defining value for determining “crowding”. If the difference in position between the current antibody and the best antibody in the current iteration is greater than the threshold \(\sigma \), crowding has not occurred and can be added to the antibody population; if the difference in position between the current antibody and the best antibody in the current iteration is less than the threshold \(\sigma \), crowding has occurred. If the difference between the current antibody and the best antibody in this iteration is less than a threshold \(\sigma \), crowding has occurred. In this case, the affinity between the two needs to be further determined, and if the current antibody has a higher affinity, this antibody can also be added to the antibody population. Otherwise, the algorithm continues to generate antibodies at random. The expression for the directed update strategy is as follows:
where M denotes the array of upper limits of antibody values, m denotes the array of lower limits of antibody values, dim denotes the antibody dimension, div denotes the distance between the current antibody and the optimal antibody during the current iteration, \(x_i^g\) denotes the i-th variable of antibody x during the g-th iteration, \(b_i^g\) denotes the i-th variable of the optimal antibody b during the g-th iteration, \(1 \le i \le \) dim, and \(g \le G_\mathrm{{max}}\).
where r denotes a uniformly distributed random number between [0, 1]. Aff(\(x_i^g\)) denotes the affinity of the i-th variable of antibody x during the g-th iteration, and Aff(\(b_i^g\)) denotes the affinity of the i-th variable of the optimal antibody b during the g-th iteration.
3.2 Improved clonal selection algorithm flow
The DUSCSA is based primarily on the clonal selection algorithm process, in which a targeted update strategy is used to determine whether an antibody is complemented or eliminated during the update phase, as shown in Fig. 5:
The implementation of the algorithm is shown in Algorithm 1:

4 Improved time complexity of the clonal selection algorithm
To predict the resources required by the algorithm and the running time of the algorithm, the time complexity of the algorithm is analyzed. The time complexity can be expressed via big O notation as O(f(n)), where f(n) is the dominant term of the complexity function. The DUSCSA contains the following main parameters: the population size D, the dimension of the antibodies dim, the clone size c, and the number of antibodies with the best affinity N. The algorithm consists of five main operational steps as follows: (1) calculation of the affinity value of the antibody; (2) cloning operation; (3) mutation operation; (4) selection operation; and (5) population directed update operation. In operation step (1), the number of executions is obtained based on the population size and is therefore D. In operation (2) and operation (3), the cloning operation on the antibody population requires cN operations. Operation (4) sorts the population to select the top N outstanding antibodies using the quicksort algorithm, which has the smallest least time complexity among sorting algorithms, and the number of operations required is \({\text {clog}}_2c\). According to the definition of the update operation in this paper, operation (5) calculates the distance between the antibody and the best antibody in the current iteration process, and a minimum of d operations are required to replace the latter d antibodies. The replacement requires a comparison of the distances, and if the antibodies are judged to be crowded, the affinity between the antibody and the optimal antibody in the current iteration process also needs to be calculated. Assuming that crowding occurs for all the antibodies that are randomly added, the affinities of each need to be calculated for further comparison, requiring a maximum number of executions of ld, where l is a constant. It is known that \(D>N\ge c\) and that \(D>d\). The total number of times the DUSCSA is executed during the gth iteration, \(S_g\), should meet the following conditions:
Since the increased time complexity of the improved algorithm compared to the clonal selection algorithm is linear in magnitude, the DUSCSA has a maximum number of operations that is O(D), which is consistent with the time complexity of the traditional clonal selection algorithm, and the update strategy does not increase the algorithm time complexity.
5 Experimental and algorithmic evaluation
In this section, 10 functions from the CEC test functions for optimization algorithms are selected to test the performance of the algorithms. The experiments are divided into two main parts: the first part compares the algorithm proposed in this paper (DUSCSA) with the classical genetic algorithm (GA) and clonal selection algorithm (CSA) to verify whether the improvements proposed in this paper are effective; the second part transposes the improvements proposed in this paper to the elite clonal selection algorithm (ECSA) proposed in the literature [12] and conducts comparison experiments with the original algorithm to verify that the proposed algorithm has an enhanced optimization effect on other clonal selection improvement algorithms. The experimental steps are as follows:
Step 1: Initialize the various parameters of the algorithm, as shown in Table 2. The genetic algorithm contains operational operators, such as crossover and variation, the clonal selection algorithm contains operational operators such as variation and cloning, and the algorithm proposed in this paper adds a crowding factor to the clonal selection algorithm. The parameters of the comparison algorithm are set according to reference [19]. The parameters of the algorithm proposed in this paper are experimentally verified, and the best performance of the algorithm is obtained when the mutation rate is set to 0.5, the initial number of clones is set to 5, and the crowding degree factor is set to 0.1.
Step 2: Set up the experimental environment, with specifics as shown in Table 3. Execute the algorithms under the same conditions and record the optimal solution for each execution of the algorithm, running each iteration for 1000 rounds, 100 times;
Step 3: Analyze the experimental results. The first part of the experiment is analyzed in two ways: first, data analysis is performed for the experimental results. We compare the affinity for a specified number of iterations, where the smaller the affinity is, the higher the accuracy of the search, and we calculate the variance for each result, where the smaller the variance is, the better the convergence stability of the algorithm. Second, convergence curve analysis is performed, where we construct a plot with the number of iterations required to obtain a given affinity on the horizontal axis, and the affinity at a certain number of iterations on the vertical axis. The lower the number of iterations is, the faster the convergence, and the smaller the affinity is, the higher the accuracy of the search. In the second part of the experiment, the results are analyzed and compared with those the original algorithm. We compare the affinity and variance, with a smaller affinity and variance indicating that the improved step incorporating other improved algorithms improve the search accuracy and convergence stability.
5.1 Test functions
The test functions selected for this paper are shown in Table 4. Among them, \(f_1 \sim f_6\) are single-peaked test functions, which have the common feature of having one global minimum, but are otherwise complex functions with multiple local minima and are suitable for testing the convergence speed and convergence stability of the detection algorithms. \(f_7\sim f_{10}\) are multipeaked test functions, which have the common feature of having multiple global minima and multiple local minima, so they are suitable for testing the performance of the detection algorithms on high-dimensional complex functions.
5.2 Analysis of the effectiveness of the algorithm
To demonstrate the effectiveness of the improvements described in this paper for the update step of the clonal selection algorithm, comparative experiments are conducted with the classic GA and the CSA to test the algorithm’s optimization accuracy, convergence speed and convergence stability by means of single-peak test functions and multipeak test functions.
For the single-peak test functions, the optimal solution (Best), mean, standard deviation (SD) and variance of the GA, CSA and DUSCSA are recorded after 100 executions of each in the same test environment, and the experimental results are shown in Table 5.
For the multipeak test functions, we set the dimension to be dim = 10, dim = 50 and dim = 100, and record the optimal solution (Best), mean, standard deviation (SD) and variance. The experimental results for each dimension are shown in Tables 6, 7 and 8.
The analysis of the data in Table 5 shows that of the three algorithms, the CSA has the best search accuracy on \(f_1\), and the DUSCSA has the best search accuracy and convergence stability on \(f_2\), \(f_3\), \(f_4\), \(f_5\), and \(f_6\). The experimental variance of DUSCSA on \(f_3\), \(f_4\) and \(f_5\) is 0, and the convergence is particularly stable. From the data in Table 6, when the dimension is 10, the GA has the best optimization accuracy on \(f_9\), the CSA has the best optimization accuracy on \(f_{10}\), and the DUSCSA has the best convergence stability on \(f_7\). In the experimental results of \(f_8\), the search accuracy of the DUSCSA is better than those of the GA and the CSA. From the data in Table 7, when the dimension is 50, the GA has the best search accuracy on \(f_8\), the CSA has the best search accuracy on \(f_9\) and \(f_{10}\), and the DUSCSA has the best accuracy and still maintains good convergence stability on \(f_7\). From the data in Table 8, the optimization accuracy of the GA is the on \(f_9\), the optimization accuracy of the CSA is the best on \(f_{10}\), and the optimization accuracy of the DUSCSA is the best on \(f_7\) and \(f_8\). Compared with the GA and the CSA, the DUSCSA has better convergence stability when the dimension is 100. Convergence graphs provide a more intuitive picture of each algorithm’s performance. Figures 6, 7, 8, 9, 10 and 11 show the convergence curves of the DUSCSA compared to those of the CSA for the six single-peak test functions.
For the single-peak test functions, the DUSCSA has more stable and reliable convergence and better accuracy than the GA and CSA. For the multipeak test functions, the optimization accuracy of all three algorithms decreases as the dimensionality increases, but the DUSCSA’s optimization accuracy is less affected by the dimensionality and maintains good convergence stability. The experimental results show that the update strategy proposed in this paper is effective, and the algorithm has good robustness and ensures stable convergence. The comparative graphs of the convergence curves of the six tested functions clearly show that the DUSCSA is more accurate than the CSA under the same number of iterations; the DUSCSA has fewer iterations than the CSA under the same affinity and therefore converges faster. The comparison of the results reveals that the overall performance of the DUSCSA is 1% higher than that of the CSA and 2.2% higher than that of the GA. In summary, the update strategy proposed in this paper is an effective way to improve the algorithm’s search accuracy and convergence stability.
5.3 Analysis of the portability of the algorithm
To demonstrate that the improvements to the update step can be used in other clonal selection improvement algorithms, the improvements in the update phase were fused into the ECSA. We compared this fused algorithm with the ordinary ECSA and recorded the optimal solution (Best), mean, standard deviation (SD) and variance after 100 executions of each in the same test environment. The experimental results are shown in Table 9.
The experimental results for \(f_1\) show that the ECSA has the higher accuracy, but among the remaining test functions, the ECSA + DUSCSA has better accuracy and convergence stability than the original algorithm. Among these functions, the experimental variance of \(f_3\), \(f_4\) and \(f_5\) is 0, and the convergence is particularly stable. The overall performance of the ECSA + DUSCSA is better than that of the ECSA by 0.97%, indicating that the improvements proposed in this paper can be transferred to other clone selection improvement algorithms and further enhance their search accuracy and convergence stability.
6 Conclusion
Many clonal selection algorithms have been studied, and improvements have been made to the cloning, mutation, and selection steps, but there is a lack of research on optimizing the update operation. In this paper, a clonal selection algorithm based on a directional update strategy is proposed to address the blindness and uncertainty of the random update of the clonal selection algorithm, which is responsible for a high number of iterations, low optimization accuracy, and poor convergence stability. If the distance is less than the threshold and the affinity is worse than that of the optimal antibody in the current iteration, then the antibody is readded to increase the probability of finding the global optimal solution and improve the convergence speed and stability of the algorithm. If the randomly added antibodies are located close to the global optimum or have high affinity, the probability of obtaining the global optimum is higher, and convergence is faster. By guiding the randomly added antibodies toward the global optimum, the algorithm converges steadily and quickly, and the accuracy of the search is higher. The evolution of the antibodies toward the global optimum is more in line with the nature of biological evolution.
The experiments demonstrate that the improvement proposed in this paper is an effective updating strategy that improves the accuracy, enhances the convergence stability and increases the convergence speed of the clonal selection algorithm. The strategy actually uses the Euclidean distance formula to calculate the similarity for determination, and more methods for calculating the similarity can be chosen for different comparison experiments to select an optimal way for improvement. The improvement proposed in this paper not only addresses the lack of research on the update phase of the clonal selection algorithm but can also be applied to other heuristic algorithms that need to perform population updates, further enhancing their optimization accuracy and convergence stability.
Availability of data and materials
This article uses 10 optimization functions from the CEC benchmark functions.
Code availability
Relevant codes can be obtained by contacting cercischinensis@foxmail.com.
References
Yan X, Li P, Tang K, Gao L, Wang L (2020) Clonal selection based intelligent parameter inversion algorithm for prestack seismic data. Inf Sci 517:86–99. https://doi.org/10.1016/j.ins.2019.12.083
Wang D, Liang Y, Dong H, Tan C, Xiao Z, Liu S (2022) Innate immune memory and its application to artificial immune systems. J Supercomput 78:1–22. https://doi.org/10.1007/s11227-021-04295-1
Chawla VK, Chanda AK, Angra S (2019) A clonal selection algorithm for minimizing distance travel and back tracking of automatic guided vehicles in flexible manufacturing system. J Inst Eng (India) Ser C 100:401–410
Yanming QUANHY (2021) Research on clonal selection algorithm for multi-robot task allocation and scheduling. J S China Univ Technol (Nat Sci Edn) 49(5):102–110
Cheikhrouhou O, Khoufi I (2021) A comprehensive survey on the multiple traveling salesman problem: applications, approaches and taxonomy. Comput Sci Rev 40:100369. https://doi.org/10.1016/j.cosrev.2021.100369
Yu-e G (2017) Research on fault recovery of ship distribution network based on cloning algorithm. Ship Sci Technol 39(16):163–165
Zhang FB, Fan XL (2018) Research on multi-objective optimal clone selection algorithm for immune intrusion detection. Comput Eng Sci 40(02):261–267
Chen Q, Fu CQP (2022) Parameter identification of permanent magnet synchronous motor based on clone-selective differential evolutionary algorithm. Sens Microsyst 41(01): 135–137141. https://doi.org/10.13873/J.1000-9787(2022)01-0135-03
Luo W, Lin X (2018) Recent advances in clonal selection algorithms and applications. Comput Intell 1:12–2. https://doi.org/10.1109/SSCI.2017.8285340
Song D, Fan X, Wen Z, Huang D, Qu X (2017) Double clonal selection algorithm based on fuzzy non-genetic information memory. J Electron Inf 39(02):255–262. https://doi.org/10.11999/JEIT160359
Li L, Lin Q, Li K, Ming Z (2021) Vertical distance-based clonal selection mechanism for the multiobjective immune algorithm. Swarm Evolut Comput 63:100886. https://doi.org/10.1016/j.swevo.2021.100886
Gálvez A, Iglesias A, Avila A, Otero C, Arias R, Manchado C (2015) Elitist clonal selection algorithm for optimal choice of free knots in b-spline data fitting. Appl Soft Comput 26:90–106. https://doi.org/10.1016/j.asoc.2014.09.030
Wanf LL, Shen Y, Xu YS et al (2021) A clone selection algorithm incorporating cloud models and backward learning. Comput Eng Appl 57(17):68–74
Wang Y, Li T, Liu X, Yao J (2022) An adaptive clonal selection algorithm with multiple differential evolution strategies. Inf Sci 604:142–169. https://doi.org/10.1016/j.ins.2022.04.043
Shu WN, Ding LX (2016) Optimization and quality factors of clone selection algorithms. J Softw 27(11):2763–2776. https://doi.org/10.13328/j.cnki.jos.004911
Tan Z, Tang Y, Li K, Huang H, Luo S (2022) Differential evolution with hybrid parameters and mutation strategies based on reinforcement learning. Swarm Evol Comput 75:101194. https://doi.org/10.1016/j.swevo.2022.101194
Zhang R-C, Pan C-Y, Wu X, Yang K (2021) Adaptive sampling immune optimization algorithm for nonlinear multi-objective probabilistic optimization problems. Acta Electronica Sinica 49(04):647–660. https://doi.org/10.12263/DZXB.20200171
Liu J-S, Ma Y-X, Li Y (2021) Improved butterfly algorithm for multi-dimensional complex function optimization problem. Acta Electronica Sinica 49(06):1068–1076
Yang C, Chen B-Q, Jia L, Wen H-Y (2020) Improved clonal selection algorithm based on biological forgetting mechanism. CompLex 2020:2807056–1280705610
Funding
This work is partially supported by the National Natural Science Foundation of China (61977021) and the Hubei Provincial Key R&D Program Project (2021BAA184).
Author information
Authors and Affiliations
Contributions
CY proposed the main structure of the algorithm in this paper, evaluated its feasibility, and proposed the flow of the algorithm in this paper. ZH performed the coding of algorithms, the collation of test functions, and the experiments of related algorithms. BJ participated in the feasibility demonstration of the algorithm and helped perform optimization of the algorithm. MZ helped analyze the algorithm time complexity and provided experimental methods. AL made great contributions in the experimental stage, including debugging after code writing and optimizing the code so that the algorithm could obtain results faster. JH helped organize the experimental datasets and record the experimental results during the experimental stage and participated in the analysis stage of the final experimental results.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or nonfinancial interests to disclose.
Ethical approval
All experimental data in this paper are real and valid, and there is no case of multiple submissions for one manuscript in this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, C., Huang, Z., Jiang, B. et al. Improved clonal selection algorithm based on the directional update strategy. J Supercomput 79, 19312–19331 (2023). https://doi.org/10.1007/s11227-023-05405-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-023-05405-x