Abstract
In this study, we propose a new genetic algorithm that uses a statistical-based chromosome replacement strategy determined by the empirical distribution of the objective function values. The proposed genetic algorithm is further used in the training process of a multiplicative neuron model artificial neural network. The objective function value for the genetic algorithm is the root mean square error of the multiplicative neuron model artificial neural network prediction. This combination of methods is proposed for a particular type of problems, that is, time-series prediction. We use different subsets of three stock exchange time series to test the performance of the proposed method and compare it against similar approaches, and the results prove that the proposed genetic algorithm for the multiplicative neuron model of the artificial neural network works better than many other artificial intelligence optimization methods. The ranks of the proposed method are 1.78 for the Nikkei data sets, 1.55 for the S&P500 data sets and 1.22 for the DOW JONES data sets for data corresponding to different years, according to the root mean square error, respectively. Moreover, the overall mean rank is 1.50 for the proposed method. Also, the proposed method obtains the best performance overall as well as the best performance for all the individual tests. The results certify that our method is robust and efficient for the task investigated.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Genetic algorithms (GAs), proposed by J. Holland, are one of the most important classes of evolutionary computation methods. GA mimics the evolutionary process of nature, based on Darwin's theory of "survival of the fittest." GAs have five basic steps: generation of initial population, selection of parents for the next generation, crossover, mutation and replacement. The initial population consisting of chromosomes corresponding to solution set is generated randomly. This is followed by selection of parents that are recombined to create offspring for the next generation. The popular selection methods in GA are roulette wheel selection, stochastic universal sampling, tournament selection, rank selection, etc. Crossover and mutation are the operators that provide diversity to the solutions in GA. The final step in GA is the replacement step. The replacement technique is used to decide which individuals (from the parent and offspring sets) will form the population of the next generation, selection that influences the algorithm convergence [35]. All these steps are important for the GA process. Parameters such as population size, crossover rate, mutation rate and the chromosomes to be eliminated (replaced) also affect the performance of the GA. In GA literature, although there are certain parameter values for the population size, crossover rate, mutation rate, there is no indicative parameter value for the chromosomes to be eliminated in the replacement step. There are various strategies that try to preserve diversity in the population while, at the same time, trying to ensure a very good convergence.
This process is known as exploration/exploitation trade-off and is a challenging aspect of evolutionary computation, with no best method known so far. In this paper, the chromosomes to be eliminated in the replacement step are determined by a statistical evaluation based on the empirical distribution of the objective function values, which is the root mean square error (RMSE). If the distribution of the RMSE values of the chromosomes of a population is the normal distribution, the mean statistics is determined as a suitable measure of central tendency. Thus, the chromosomes with RMSE values greater than the mean statistic are eliminated in the replacement step. Otherwise, if the distribution of the RMSE values is not normal, the median statistic is determined as a suitable measure of central tendency for the objective function values. Thus, the chromosomes with RMSE values greater than the median statistic are eliminated in the replacement step. A new approach called statistical-based replacement is proposed to determine the chromosomes to be eliminated in the replacement. The number of chromosomes to be eliminated is determined automatically, preserving about 50% of the chromosomes from the parents and offspring population. The performance of the proposed GA approach is evaluated on the training process of single multiplicative neuron model artificial neural networks (SMNM-ANN) proposed in [30].
The paper is organized as follows: in Sect. 2, we introduce the statistical-based replacement method. In Sect. 3, we give a brief introduction to SMNM-ANN. Section 4 summarizes very briefly the GA. Our proposed methodology is given in Sect. 5. Section 6 compares the results of our proposed algorithm against other methods. Finally, the paper ends with a discussion and conclusions in Sect. 7.
2 The proposed statistical-based replacement procedure
Determining the number of chromosomes to be eliminated is a very important step in the GA iterative process. The size of the population usually remains constant at each iteration. This is achieved in the replacement phase, where a constant number from the parent and offspring population is preserved for the next generation. Replacement is the last step of any genetic algorithm cycle.
Let us consider the objective function values obtained for each chromosome in the population as a data set. We verify whether this data set has a normal distribution or not. The Lilliefors test is used to check the normality of the distribution. When a data set has a normal distribution, the mean statistic is the best measure of central tendency. If a data set does not have a normal distribution, the median statistic can be preferred as a measure of central tendency.
From this statistical perspective, if the relevant data set (the objective function values of each chromosome in our case) has a normal distribution, the chromosomes with an objective function value greater than the mean statistics (obtained by dividing the sum of the objective function values of each chromosome in the population to the total number of chromosomes) are eliminated for an objective function that requires minimization. When the objective function needs to be maximized, the chromosomes with an objective function value smaller than the mean statistics are eliminated.
Otherwise, if the relevant data set does not have a normal distribution, the chromosomes with an objective function value greater than the median statistics are eliminated, in the case of objective minimization. In case of objective function maximization, the chromosomes with an objective function value smaller than the median statistics are eliminated. This strategy is called as a statistical-based replacement in this paper.
An example of this representation is given in Fig. 1. There are \(n\) chromosomes, and each chromosome has \(g\) genes. OFV shows the objective function value obtained for each chromosome, and the median statistic of OFV is calculated by Eq. (1):
where \(X\) represents the median rank of the sorted OFV values.
3 Single multiplicative neuron model artificial neural networks
SMNM-ANN proposed in [30] is an artificial neural network that uses multiplicative neuron in its structure instead of additive neuron (that is most common with ANNs models). The authors use backpropagation learning (BP) algorithm in the training of SMNM-ANN.
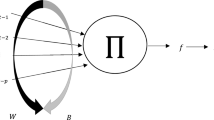
The structure of SMNM-ANN is given in Fig. 2.
In Fig. 2, f represents the activation function, the join function \(\Omega\) is multiplication, the weight and bias vector is \(\Theta =({w}_{1}, {w}_{2},\ldots ,{w}_{m},{b}_{1}, {b}_{2},\ldots ,{b}_{m})\) and the inputs are \(({x}_{1}, {x}_{2},\ldots ,{x}_{m})\). The neuron model with \(m\) input variables \(\left( {x_{i} , i = 1,2, \ldots ,m} \right)\) given in Fig. 2 has \(\left( {2 \times m} \right)\) elements. Of these, \(m\) correspond to the weights \(\left( {w_{i} , i = 1,2, \ldots ,m} \right)\) and the other \(m\) to the biases \(\left( {b_{i} , i = 1,2, \ldots ,m} \right)\). The output of the SMNM-ANN with the logistic activation function is calculated using Eqs. (2) and (3):
Many heuristic optimization algorithms have also been used in the training process of SMNM-ANN in recent years. [24] uses particle swarm optimization (PSO), [22] uses improved back propagation algorithm, [6] uses harmony search algorithm, [37] employs online training algorithms, [19] uses improved glow-worm swarm optimization algorithm, [10] uses differential evolution algorithm (DEA), [11] uses artificial bat algorithm (ABA) for the optimization of SMNM-ANN. [12] also used PSO for the training of SMNM-ANN and proposed a robust learning algorithm for SMNM-AN. [38] makes use of SMNM-ANN for forecasting of hourly wind speed. [27] used SMNM-ANN based on Gaussian activation function. [28] proposed a SMNM-ANN with autoregressive coefficient for time-series modeling. [13] uses sine cosine algorithm and [9] uses a hybrid ABA and BP algorithm in the training process of SMNM-ANN. [4] proposes a SMNM-ANN based on threshold for non-linear time-series forecasting.
4 Genetic algorithms
There are many metaheuristics optimization algorithms, with different properties, available in the literature. Among these algorithms, harmony search algorithm aims to obtain the best harmony from different melodies, sine cosine optimization algorithm employs the properties of sine and cosine functions, symbiotic learning algorithm is linked to the optimization process of symbiotic relationships in nature, and GA is a population-based heuristic optimization algorithm [18]. Many popular metaheuristics optimization algorithms similar to these are clearly explained, together with the algorithm provided, in the studies of [16, 36] and [23].
Among these methods, GA is one of the oldest known heuristic algorithms. GA differs from other heuristic optimization algorithms with its unique paradigm and operators used in the optimization process.
GA is a search and optimization method inspired from natural selection and genetics. GA is an approximation method, generally very effective in searching over very large (almost intractable) search space and in finding a good solution. The elements of a GA are called chromosomes, the basic structural units of the chromosome are called genes, and the values of genes are alleles. To solve a problem with a GA, it is necessary to create an initial population, to determine the types and rates of crossover and mutation operators, the selection operator and the stopping criterion. All of these parameters and operators greatly affect the overall performance.
The chromosomes correspond to potential solutions. If the GA has a good initial population, the algorithm is considered to have a better chance of finding a good solution. The population size parameter, which is generally defined by the user, is one of the important factors affecting the performance of the GA. It is known that small population size can direct the algorithm to non-optimal solutions, and large population size may require more computation time for the algorithm to find a solution. The crossover operator, one of the most basic operators in GA, is an operator that allows the creation of new chromosomes according to a predetermined recombination mechanism applied to selected chromosome pairs. The mutation operator is an operator applied generally with a lower probability. The mutation operator takes a random walk around the chromosomes and has the effect of replacing the allele of a random gene of a randomly selected chromosome. In the final step of GA, which is the replacement of the population for the next generation, the chromosomes from the old population are replaced by new ones.
This process is iterative, repeated until a stopping condition is met.
GA has been used for different aims with ANNs. [17and [29] proposed a method based on GA and ANN. GA-based ANN has been used for scheduling problems in [1, 2], fault diagnosis model in [33], for bankruptcy prediction in [5], and for forecasting in [34]. GA has been used for designing ANN in [3], [14] and [26]. [8] and [32] trained the feed-forward ANN with GA. [25] used GA for missing data problems in ANN. [20] and [31] compared GA with backpropagation and simulated annealing, respectively. [21] compared GA-based ANN with a statistical methods. [39] used GA for input selection in ANN.
5 The proposed genetic algorithm with statistical-based replacement procedure for the training of SMNM-ANN
Determining the size and the elements of the chromosome set to be eliminated in the replacement step appears to be a problem in the GA process. In this paper, we propose a method to overcome the problem of selecting the chromosomes to be eliminated in the replacement step. We use a statistical evaluation that takes into account a statistical-based replacement procedure. How many and what chromosomes to be eliminated is determined by whether the RMSE value of a chromosome is greater or smaller than the mean or median of the objective function values of the entire population. Chromosomes with values greater than the mean or median RMSE values of all chromosomes in the population are eliminated. The innovations and advantages of the proposed method are as follows:
-
The method is systematic and statistically based;
-
Important genes are preserved due to the fact that the top 50% of the chromosomes in the population are not eliminated;
-
The convergence speed of GA is increased by using the greedy selection strategy in the mutation and crossover steps;
-
The restart strategy is used to increase the probability that the algorithm will escape the local optimum trap;
-
In order to prevent the overfitting problem of the artificial neural network, the number of iterations in which the genetic algorithm cannot provide improvement in the best chromosome of the population is checked as an early stop condition.
The steps of the proposed methodology are given below.
Step 1. Set GA and SMNM-ANN parameters.
These parameters are:
-
\(nc\): the number of chromosomes in the population;
-
\(ng\): the number of genes in a chromosome;
-
\(cor\): the crossover ratio;
-
\(mr\): the mutation ratio;
-
\(m\): the number of the inputs for the network;
-
n: the size of the training set.
Step 2. Generate the initial population.
The initial population is generated by considering the parameters \(nc, ng {\text{ and }} m\). Since an SMNM-ANN with \(m\) input will have \(m\) bias and \(m\) weight values, each chromosome in the population has a total of \(\left( {2 \times m} \right)\) genes. These gene values are generated with a uniform distribution between zero and one \(\left( {U\left( {0,1} \right)} \right)\). A population structure with \(m\) and \(nc\) parameters is represented in Fig. 3.
Step 3. Calculate the objective function values of each chromosome in the population.
For this aim, the net values of each chromosome are first calculated using Eqs. (2 and 3). Then, these net values are used for the calculation of SMNM-ANN output ( \(\hat{y}_{t}\)). Finally, the RMSE criterion given in Eq. (4) is used as the fitness function value.
Step 4. Apply the genetic operators.
Step 4.1. Apply crossover operator.
Before applying the crossover operator, the number of chromosome pairs to be crossed is determined by the \(cor\) parameter. The chromosomes are selected randomly from the population and the mutual genes of chromosome pairs to be crossed are displaced. The new chromosome obtained as a result is evaluated according to the greedy selection strategy and is included in the population if it has a fitness value better than the old chromosome. Otherwise, the old chromosome is stored in the population.
Step 4.2. Apply the mutation operator.
The number of chromosomes to be mutated is determined by the multiplication of \(nc\) and \(mr\) parameters. The chromosomes are selected randomly from the population and the gene/genes to be changed are also randomly selected. The new chromosome obtained as a result of the mutation is evaluated according to the greedy selection strategy and is included in the population if it has a better fitness value than the parent chromosome. Otherwise, the old chromosome is stored in the population.
Step 5. Apply the restart strategy.
The number of iterations is checked. If a pre-set number of iterations is reached (this is set to 200 in our work), all chromosomes in the population are randomly reproduced.
Step 6. Determine the number of chromosomes to be eliminated.
After calculating all RMSE values corresponding to each chromosome in the population, these RMSE values are subjected to normality test using Lilliefors test. If the RMSE values have a normal distribution, calculate the mean of these RMSE (meanRMSE) values and the RMSE values corresponding to chromosomes greater than meanRMSE are eliminated. If these RMSE values do not have normal distribution, calculate the median of these RMSE (medianRMSE). The chromosomes whose RMSE values are greater than medianRMSE are eliminated. The eliminated chromosomes are replaced by new generated chromosomes.
Step 7. Check the stopping condition.
If the maximum number of iterations or a number of consecutive iterations with no improvements has been reached, the process is ended. Otherwise, go to Step 3. The flow chart is given for the algorithm of the proposed method in Fig. 4.
6 Experiments
In the application part of the paper, a detailed analysis is carried out to evaluate the performance of the proposed method. S&P500, DOW JONES, and NIKKEI stock exchange closing time-series data sets between 2016 and 2018 are analyzed with SMNM-ANN trained by particle swarm optimization algorithm (PSO) proposed in [24], SMNM-ANN trained by artificial bat algorithm (BAT) proposed by [11], SMNM-ANN trained by a hybrid of artificial bat algorithm and backpropagation algorithms (BAT-BP) proposed by [9], SMNM-ANN trained by differential evolution algorithm (DEA) proposed by [10], SMNM-ANN trained by GA, SMNM-ANN trained by artificial bee colony algorithm (ABC), SMNM-ANN trained by backpropagation algorithm (BP) proposed by [30].
The performance of each time series analyzed is tested on different test set lengths. For this purpose, the test set lengths for each year of each time series are taken as 10, 20, and 50. In the use of the proposed SBGA, the number of inputs varies between 1 to 5 with an increment of 1, the number of chromosomes in the population varies between 30 and 100 with an increment 10, the crossover rate takes values between 0.1 and 0.7 with an increment 0.1, the mutation rate values used are 0.01, 0.05, 0,1, 0.15 and 0.2, and the parameter sets for the best result for each year of each time series with different test lengths are given in Table 1. The parameter settings used for the proposed method are the same as the ones used by the other methods we compared our results with.
An evaluation of all the methods analyzed in this paper is done considering the RMSE metric (given in Eq. (4)) and mean absolute percentage error (MAPE) criteria given in Eq. (5):
The RMSE and MAPE results for the S&P500 stock exchange time-series test data for different years are given in Table 2. When analyzing the results, we can observe that the proposed SBGA method is the best in terms of RMSE criterion when the length test is 20 and 50 for S&P500 2016 test data set and for all the test lengths (10, 20, and 50) for SP500 2018 test data set.
SBGA method also obtains the best results for MAPE criterion when the length test is 10 for S&P500 2017 test data set. Results are also graphically given in Fig. 5.
Overall, the results are also very good for the other values of the parameters.
We next look at the DOW JONES stock exchange data sets. The RMSE and MAPE results are provided in Table 3. From the results presented, we can observe that SBGA method is the best in terms of RMSE and MAPE criteria when the length test is 10 and 20 for DOW JONES 2016 test data set. SBGA method obtains the best results for.
RMSE criterion and the second-best values for MAPE metric when the length test is 50 for DOW JONES 2017 test data set. Also, SBGA method is the best in terms of RMSE criteria when the length test is 10 and 50 for DOW JONES 2017 test data set and the length test is 10, 20 and 50 for DOW JONES 2018 test data set. Results are given in Fig. 6.
The last experiment performed is for NIKKEI data set, with the results presented in Table 4. SBGA method is the best in terms of RMSE criterion when the length test is 20 and 50 for NIKKEI 2016 test data set and when the length test is 10, 20, and 50 for NIKKEI 2018 test data set. SBGA method obtains the best results for the MAPE criterion when the length test is 10 for NIKKEI 2017 test data set. Results are also graphically given in Fig. 7.
Finally, we rank all the methods compared in this study in terms of RMSE results for each of the parameter configuration considered. Results are presented in Table 5.
7 Conclusions and discussions
In the GA literature, although there are certain parameter values for population size, crossover and mutation rates, there is no specific value or percentage for the number of chromosomes to be eliminated in the replacement step. In order to fill this gap, a new replacement operator named statistical-based replacement procedure based on the distribution of the objective function values is proposed in this paper. The new method is a statistical-based replacement procedure and uses the mean or median statistics through the replacement procedure, regardless of whether the value of the objective function has a normal distribution or not. The proposed statistical-based replacement procedure is used in the training process of SMNM-ANN. The performance of the proposed method is assessed on different test sets with different test lengths. The analysis results show that the proposed method is the best among all the methods used. It can be also said that the GA method using statistical-based replacement has a significantly superior performance compared to the classical genetic algorithm.
For each data set analysis, the rank numbers are computed for all methods. The mean of the rank numbers is calculated by using ranks as sample data for different test set lengths. The mean rank numbers of the proposed method are 1.83, 2.83 and 1.5 for the Nikkei data sets, 3.16, 2 and 1.66 for the S&P500 data sets and 1.33, 1.83 and 1.66 for the DOW JONES data sets for different years, respectively. The proposed method is the best or second-best method for all individual analyses, and it has the best performance overall.
In future studies, the proposed method can be used for the training of different artificial neural network types and in other application areas with similar data types.
Data availability
The data used in this work are publicly available in open repositories.
References
Delgoshaei A, Aram A, Mantegh V, Hanjani S, Nasiri AH, Shirmohamadi F (2019) A multi-objectives weighting genetic algorithm for scheduling resource-constraint project problem in the presence of resource uncertainty. Int J Supply Op Manag 6(3):213–230
Delgoshaei A, Ariffin M, Baharudin BHTB, Leman Z (2015) minimizing makespan of a resource-constrained scheduling problem: a hybrid greedy and genetic algorithm. Int J Ind Eng Comput 6(4):503–520
Jones AJ (1993) Genetic algorithms and their applications to the design of neural networks. Neural Comput Appl 1(1):32–45
Yildirim AN, Bas E, Egrioglu E (2021) Threshold single multiplicative neuron artificial neural networks for non-linear time series forecasting. J Appl Stat 48(13–15):2809–2825
Back B, Laitinen T, Sere K (1996) Neural networks and genetic algorithms for bankruptcy predictions. Expert Syst Appl 11(4):407–413
Worasucheep C (2012) Training a single multiplicative neuron with a harmony search algorithm for prediction of S&P500 index-An extensive performance evaluation, In: Knowl Smart Technol (KST), IEEE, pp 1–5.
Whitley D (1995) Genetic algorithms and neural networks, genetic algorithms in engineering and computer. Science 3:203–216
Montana DJ, Davis L (1989) Training feedforward neural networks using genetic algorithms. In IJCAI 89:762–767
Bas E, Egrioglu E, Yolcu U (2020) A hybrid algorithm based on artificial bat and backpropagation algorithms for multiplicative neuron model artificial neural networks. J Ambient Intell Human Comput. https://doi.org/10.1007/s12652-020-01950-y
Bas E (2016) The training of multiplicative neuron model based artificial neural networks with differential evolution algorithm for forecasting. J Artif Intell Soft Comput Res 6(1):5–11
Bas E, Yolcu U, Egrioglu E, Yolcu OC, Dalar AZ (2016) Single multiplicative neuron model artificial neuron network trained by bat algorithm for time series forecasting. Am J Intell Syst 6(3):74–77
Bas E, Uslu VR, Egrioglu E (2016) Robust learning algorithm for multiplicative neuron model artificial neural networks. Expert Syst Appl 56:80–88
Kolay E (2019) A novel multiplicative neuron model based on sine cosine algorithm for time series prediction. Eskişehir Tech Univ J Sci Technol A-Appl Scie Eng 20(2):153–160
Miller GF, Todd PM, Hegde SU (1989) Designing neural networks using genetic algorithms. In ICGA 89:379–384
Lilliefors HW (1967) On the Kolmogorov-Smirnov test for the exponential distribution with mean unknown. J Am Stat Assoc 64:387–389
Boussaïd I, Lepagnot J, Siarry P (2013) A survey on optimization metaheuristics. Inf Sci 237:82–117
Schaffer JD, Whitley D, Eshelman LJ (1992) Combinations of genetic algorithms and neural networks: a survey of the state of the art, In [Proceedings] COGANN-92: international Workshop on Combinations of Genetic Algorithms and Neural Networks, IEEE, pp 1–37
Holland JH (1975) Adaptation in natural and artificial systems: an introductory analysis with applications to biology, control, and artificial intelligence. U Michigan Press
Cui JHH, Feng J, Guo J, Wang T (2015) A novel single multiplicative neuron model trained by an improved glowworm swarm optimization algorithm for time series prediction. Knowledge 88:195–209
Gupta JN, Sexton RS (1999) Comparing backpropagation with a genetic algorithm for neural network training. Omega 27(6):679–684
Hansen JV, McDonald JB, Nelson RD (1999) Time series prediction with genetic-algorithm designed neural networks: an empirical comparison with modern statistical models. Comput Intell 15(3):171–184
Burse K, Manoria M, Kirar VPS, (2011) Improved back propagation algorithm to avoid local minima in multiplicative neuron model, In: International Conference on Advances in Information Technology and Mobile Communication, Springer, Berlin, Heidelberg pp 67–73.
Hussain K, Salleh MNM, Cheng S, Shi Y (2019) Metaheuristic research: a comprehensive survey. Artif Intell Rev 52(4):2191–2233
Zhao L, Yang Y (2009) PSO-based single multiplicative neuron model for time series prediction. Expert Syst Appl 36(2):2805–2812
Abdella M, Marwala T (2005) The use of genetic algorithms and neural networks to approximate missing data in database, In IEEE 3rd International Conference on Computational Cybernetics, IEEE, pp 207–212
Idrissi MAJ, Ramchoun H, Ghanou Y, Ettaouil M (2016) Genetic algorithm for neural network architecture optimization, In: 2016 3rd International Conference on Logistics Operations Management (GOL), IEEE, pp 1–4
Gundogdu O, Egrioglu E, Aladag CH, Yolcu U (2016) Multiplicative neuron model artificial neural network based on Gaussian activation function. Neural Comput Appl 27(4):927–935
Yolcu OC, Bas E, Egrioglu E, Yolcu U (2018) Single multiplicative neuron model artificial neural network with autoregressive coefficient for time series modelling. Neural Process Lett 47(3):1133–1147
Duvigneau R, Visonneau M (2002) Hybrid genetic algorithms and neural networks for fast CFD-based design. In: 9th AIAA/ISSMO Symposium on Multidisciplinary Analysis and Optimization (p. 5465)
Yadav RN, Kalra PK, John J (2007) Time series prediction with single multiplicative neuron model. Appl Soft Comput 7(4):1157–1163
Sexton RS, Dorsey RE, Johnson JD (1999) Optimization of neural networks: a comparative analysis of the genetic algorithm and simulated annealing. Eur J Oper Res 114(3):589–601
Ding S, Su C, Yu CJ (2011) An optimizing BP neural network algorithm based on genetic algorithm. Artif intell Rev 36(2):153–162
Sampath S, Singh R (2006) An integrated fault diagnostics model using genetic algorithm and neural networks. ASME J Eng Gas Turbines Power 128(1):49–56
Ling SH, Leung FHF, Lam HK, Lee YS, Tam PKS (2003) A novel genetic-algorithm-based neural network for short-term load forecasting. IEEE Trans Ind Electron 50(4):793
Sivanandam SN, Deepa SN (2008) Genetic algorithms. In Introduction to Genetic Algorithms, Berlin
Dokeroglu T, Sevinc E, Kucukyilmaz T, Cosar A (2019) A survey on new generation metaheuristic algorithms. Comput Ind Eng 137:106040
Wu X, Mao J, Du Z, Chang Y (2013) Online training algorithms based single multiplicative neuron model for energy consumption forecasting. Energy 59:126–132
Wu X, Zhu Z, Su X, Fan S, Du Z, Chang Y, Zeng Q (2015) A study of single multiplicative neuron model with nonlinear filters for hourly wind speed prediction. Energy 88:194–201
Guo Z, Uhrig RE (1992) Using genetic algorithms to select inputs for neural networks, In: [Proceedings] COGANN-92: International Workshop on Combinations of Genetic Algorithms and Neural Networks, IEEE, pp 223–234
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
There is no declared conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Egrioglu, E., Grosan, C. & Bas, E. A new genetic algorithm method based on statistical-based replacement for the training of multiplicative neuron model artificial neural networks. J Supercomput 79, 7286–7304 (2023). https://doi.org/10.1007/s11227-022-04935-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11227-022-04935-0