Abstract
We report hydration properties of several DNA-binding ligands (pharmaceutical drug caffeine; mutagens proflavine, ethidium bromide, propidium iodide; polyamines putrescine, spermine), which are able to interact with DNA in different modes via external binding, intercalation or minor groove binding. We show that the detection of the bound water molecules and the estimation of their amount in aqueous solutions of ligands can be efficiently carried out using the measurements of complex dielectric permittivity of the solutions in the millimeter range of radio waves. Our dielectrometric data are combined with the results of the molecular modeling including quantum chemical calculations and Monte Carlo simulations. We show that number of water molecules able to form hydrogen bonds with donor–acceptor groups of ligands correlates with hydration numbers taken from the literature or obtained in EHF dielectrometry experiment. The latter indicates that the EHF dielectrometry method is sensitive for the tightly bound water molecules.





Similar content being viewed by others
References
Haq I (2002) Thermodynamics of drug–DNA interactions. Arch Biochem Biophys 403:1–15. doi:10.1016/S0003-9861(02)00202-3
David-Eden H, Mankin AS, Mandel-Gutfreund Y (2010) Structural signatures of antibiotic binding sites on the ribosome. Nucl Acids Res 38:5982–5994. doi:10.1093/nar/gkq411
Han F, Chalikian TV (2003) Hydration changes accompanying nucleic acid intercalation reactions: volumetric characterizations. J Am Chem Soc 125:7219–7229. doi:10.1021/ja030068p
Mancera RL (2007) Molecular modeling of hydration in drug design. Curr Opin Drug Discov Devel 10(3):275–280
Lane AN, Jenkins TC (2000) Thermodynamics of nucleic acids and their interactions with ligands. Q Rev Biophys 33(3):255–306
Shi X, Macgregor RB Jr (2007) Volume and hydration changes of DNA–ligand interactions. Biophys Chem 125:471–482. doi:10.1016/j.bpc.2006.10.011
Marky LA, Kupke DW, Kankia BI (2001) Volume changes accompanying interaction of ligands with nucleic acids. Meth Enzymol 340:149–165. doi:10.1016/S0076-6879(01)40421-6
Thomas JR, Hergenrother PJ (2008) Targeting RNA with small molecules. Chem Rev 108:1172–1224. doi:10.1021/cr0681546
Baron JJ, Roberts H (1981) Caffeine. Springer, Berlin
Dusenbery DV, Uretz RB (1972) The interaction of acridine dyes with the densely packed DNA of bacteriophage. Biophys J 12:1056–1065. doi:10.1016/S0006-3495(72)86143-5
Girousi STh, Alexaidou DK, Ioannou AK (2008) An electroanalytical study of the drug proflavine. Microchim Acta 160:435–439. doi:10.1007/s00604-007-0812-1
Finkelstein T, Weinstein IB (1967) Proflavine binding to transfer ribonucleic acid, synthetic ribonucleic acids and deoxyribonucleic acid. J Biol Chem 242(17):3763–3768
Neidle S, Pearl LH, Herzyk P, Berman HM (1988) A molecular model for proflavine-DNA intercalation. Nucl Acids Res 16:8999–9016. doi:10.1093/nar/16.18.8999
Huang Q, Fu WL (2005) Comparative analysis of the DNA staining efficiencies of different fluorescent dyes in preparative agarose gel electrophoresis. Clin Chem Lab Med 43:841–842. doi:10.1515/CCLM.2005.141
Waring MJ (1965) Complex formation between ethidium bromide and nucleic acids. J Mol Biol 13:269–282. doi:10.1016/S0022-2836(65)80096-1
Lecoeur H (2002) Nuclear apoptosis detection by flow cytometry: influence of endogenous endonucleases. Exp Cell Res 277(1):1–14. doi:10.1006/excr.2002.5537
Stevenson P, Sones KR, Gicheru MM, Mwangi EK (1995) Comparison of isometamidium chloride and homidium bromide as prophylactic drugs for trypanosomiasis in cattle at Nguruman, Kenya. Acta Trop 59:257–258. doi:10.1016/0001-706X(94)00080-K
Lawrence SA (2004) Amines: synthesis, properties and applications. Cambridge University Press, Cambridge. ISBN 978-0-521-78284-5
Morrison JC, Filer CN (2015) Tritiation and characterization of several polyamine natural products. Appl Radiat Isot 98:71–73. doi:10.1016/j.apradiso.2015.01.017
Zhang L, Zhao M, Yuan JL (2013) Related research progress on Alzheimer’s disease and its microecological control. Zhongguo Weishengtaixue Zazhi 25:861–865
Battaglia V, De Stefano Shields C, Murray-Stewart T, Casero RA (2014) Polyamine catabolism in carcinogenesis: potential targets for chemotherapy and chemo-prevention. Amino Acids 46:511–519. doi:10.1007/s00726-013-1529-6
Mendez JD, Palomar-Morales M (1999) Preventionby l-arginine and polyamines of delayed development and embryo toxicity caused by chemically-induced diabetes in rats. Reprod Toxicol 13:501–509. doi:10.1016/S0890-6238(99)00039-8
Liquori AM, Constantino L, Crescenzi V, Elia V, Giglio E, Puliti R, De Santis Savino M, Vitagliano V (1967) Complexes between DNA and polyamines: a molecular model. J Mol Biol 24:113–122. doi:10.1016/0022-2836(67)90094-0
Vijayanathan V, Thomas T, Antony T (2004) Formation of DNA nanoparticles in the presence of novel polyamine analogues: a laser light scattering and atomic force microscopic study. Nucl Acids Res 32:127–134. doi:10.1093/nar/gkg936
Rider JE, Hacker A, Mackintosh CA, Pegg AE, Woster PM, Casero RA Jr (2007) Spermine and spermidine mediate protection against oxidative damage caused by hydrogen peroxide. Amino Acids 33:231–240. doi:10.1007/s00726-007-0513-4
Kashpur VA, Maleev VYA, Shchegoleva TYU (1976) Globular protein hydration by a differential dielectrometric method. Mol Biol 10:568–575
Kashpur VA, Maleev VYA, Khorunzhaya OV (2010) Application of differential EHF dielectrometry in molecular biophysics. Telecommun Radio Eng 69(16):1473–1489. doi:10.1615/TelecomRadEng.v69.i16.80
Craig DQM (1995) Dielectric analysis of pharmaceutical systems. Taylor & Francis, London. ISBN 0-13-210279-X
Danilov VI, Tolokh IS (1984) Nature of the stacking of nucleic acids bases in water: a Monte Carlo simulation. J Biomol Struct Dyn 2:119–130. doi:10.1080/07391102.1984.10507551
Poltev VI, Malenkov GG, Gonzalez EJ, Teplukhin AV, Rein R, Shibata M, Miller JH (1996) Modeling DNA hydration: comparison of calculated and experimental hydration properties of nucleic acid bases. J Biomol Struct Dyn 13:717–726. doi:10.1080/07391102.1996.10508884
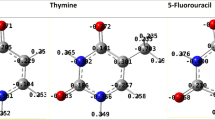
Alderfer JL, Danilov VI, Poltev VI, Slyusarchuk ON (1999) A study of the hydration of deoxydinucleoside monophosphates containing thymine, uracil and its 5-halogen derivatives: Monte Carlo simulation. J Biomol Struct Dyn 16:1107–1117. doi:10.1080/07391102.1999.10508319
Shestopalova AV (2002) Hydration of nucleic acids components in dependence of nucleotide composition and relative humidity: a Monte Carlo simulation. Eur Phys J D 20:331–337. doi:10.1140/epjd/e2002-00145-8
Maleev VYA, Semenov MA, Kashpur VA, Bolbukh TV, Shestopalova AV, Anischenko DB (2002) Structure and hydration of polycytidylic acid from the data of infrared spectroscopy, EHF dielectrometry and computer modeling. J Mol Struct 605:51–61. doi:10.1016/S0022-2860(01)00666-4
Danilov VI, Shestopalova AV (1989) Hydrophobic effect in biological associates: a Monte Carlo simulation of caffeine molecules stacking. Int J Quantum Chem 35:103–112. doi:10.1002/qua.560350105
Danilov VI, Slyusarchuk ON, Poltev VI, Alderfer JL, Wollman RM, Brickmann JA, Lautenschlager P (1992) A Monte Carlo simulation of hydration of xanthine-derivatives and their stacked forms. J Biomol Struct Dyn 9:1239–1252. doi:10.1080/07391102.1992.10507989
Maleev VYA, Semenov MA, Kruglova EB, Bolbukh TV, Gasan AI, Bereznyak EG, Shestopalova AV (2003) Spectroscopic and calorimetric study of DNA interaction with a new series of actinocin derivatives. J Mol Struct 645:145–158. doi:10.1016/S0022-2860(02)00541-0
Ellison WJ, Lamkaouchi K, Moreau JM (1996) Water: a dielectric reference. J Mol Liq 68:171–279. doi:10.1016/0167-7322(96)00926-9
Schmidt MW, Baldridge KK, Boatz JA, Elbert ST, Gordon MS, Jensen JH, Koseki S, Matsunaga N, Nguyen KA, Su S, Windus TL, Dupuis M, Montgomery JA (1993) General atomic and molecular electronic structure system. J Comput Chem 14:1347–1363. doi:10.1002/jcc.540141112
Neese F (2012) The ORCA Program system. Comput Mol Sci 2(1):73–78. doi:10.1002/wcms.81
Kostjukov VV, Khomytova NM, Hernandez Santiago AA, Licona Ibarra R, Davies DB, Evstigneev MP (2011) Calculation of the electrostatic charges and energies for intercalation of aromatic drug molecules with DNA. Int J Quantum Chem 111:711–721. doi:10.1002/qua.22451
Singh UC, Kollman PA (1984) An approach to computing electrostatic charges for molecules. J Comput Chem 5:129–145. doi:10.1002/jcc.540050204
Breneman CM, Wiberg KB (1990) Determining atom-centered monopoles from molecular electrostatic potentials. The need for high sampling density in formamide conformational analysis. J Comput Chem 11(3):361–373. doi:10.1002/jcc.540110311
Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE (2000) The Protein Data Bank. Nucleic Acids Res 28:235–242. doi:10.1093/nar/28.1.235
Metropolis N, Rosenbluth A, Rosenbluth M, Teller A, Teller E (1953) Equation of state calculations by fast computing machines. J Chem Phys 21:1087–1092. doi:10.1063/1.1699114
Poltev VI, Grokhlina TI, Malenkov GG (1984) Hydration of nucleic acid bases studied using novel atom–atom potential functions. J Biomol Struct Dyn 2:413–429. doi:10.1080/07391102.1984.10507576
Gonzalez E, Cedeno FI, Teplukhin AV, Malenkov GG, Poltev VI (2000) Refinement of methodology for simulation of nucleic acid hydration. Rev Mex Fis 46:142–147
Teplukhin AV, Malenkov GG, Poltev VI (1998) Monte Carlo simulation of DNA fragment hydration in the presence of alkaline cations using novel atom–atom potential functions. J Biomol Struct Dyn 16:289–300. doi:10.1080/07391102.1998.10508247
Poltev VI, Grokhlina TI (2002) A molecular mechanics approach to structure-activity relations in DNA-ligands. Anti-cancer drug design: biological and biophysical aspects of synthetic phenoxazone derivatives. SEVNTU Press, Sevastopol, pp 187–241
Malenkov G (2009) Liquid water and ices: understanding the structure and physical properties. J Phys: Condens Matter 21(28):283101. doi:10.1088/0953-8984/21/28/283101
Abraham FF (1974) Monte Carlo simulation of physical clusters of water molecules. J Chem Phys 61:1221–1225. doi:10.1063/1.1681997
Abraham FF (1982) Statistical surface physics: a perspective via computer simulation of microclusters, interfaces and simple films. Rep Prog Phys 45:1113–1162. doi:10.1088/0034-4885/45/10/002
Jeffrey GA, Saenger W (1991) Hydrogen bonding in biological structures. Springer, Berlin
Mashimo S, Kuwabara S, Yagihara S (1987) Dielectric relaxation time and structure of bound water in biological materials. J Phys Chem 91:6337–6338. doi:10.1021/j100309a005
Buchanan TJ, Haggis GM, Hasted JB (1952) The dielectric estimation of protein hydration. Proc R Soc A 213:379–391. doi:10.1098/rspa.1952.0132
Wei Y, Sridhar S (1992) Biological applications of a technique for broadband complex permittivity measurements. IEEE MTT-S Digest 3:1271–1274. doi:10.1109/MWSYM.1992.188233
Akhadov YY (1977) Dielectric properties of binary solutions. Nauka, Moscow
Wachter W, Kunz W, Buchner R (2005) Is there an anionic Hofmeister effect on water dynamics? Dielectric spectroscopy of aqueous solutions of NaBr, NaI, NaNO3, NaClO4, and NaSCN. J Phys Chem A 109:8675–8683. doi:10.1021/jp053299m
Shestopalova AV (2006) The investigation of the association of caffeine and actinocin derivatives in aqueous solution: a molecular dynamics simulation. J Mol Liq 127(1–3):113–117. doi:10.1016/j.molliq.2006.03.045
Tavagnacco L, Schnupf U, Mason PE, Saboungi ML, Cesàro A, Brady JW (2011) Molecular dynamics simulation studies of caffeine aggregation in aqueous solution. J Phys Chem B 115(37):10957–10966. doi:10.1021/jp2021352
Bhanita Sh, Sandip P (2013) Effects of dilute aqueous NaCl solution on caffeine aggregation. J Chem Phys 139:194504. doi:10.1063/1.4830414
Kentsis A, Mezei M, Osman R (2003) MC-PHS: A Monte Carlo implementation of the primary hydration shell for protein folding and design. Biophys J 84(2):805–815. doi:10.1016/S0006-3495(03)74900-5
Acknowledgments
The study was supported in part by the SCST Programme of Implementation and Usage of GRID technologies for 2009–2013. The calculations were carried out using clusters of Institute for Scintillation Materials NASU and Institute for Radiophysics and Electronics NASU. The research which results are presented in the present paper was fulfilled due to the collaboration with Prof. O. Shishkin. The authors also thank Roman Zubatyuk (ISMA NASU) for the assistance in quantum chemical calculations.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shestopalova, A.V., Pesina, D.A., Kashpur, V.A. et al. Hydration of DNA-binding biological active compounds: EHF dielectrometry and molecular modeling results. Struct Chem 27, 159–173 (2016). https://doi.org/10.1007/s11224-015-0695-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-015-0695-4