Abstract
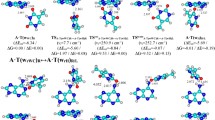
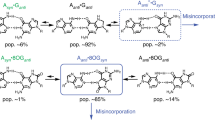
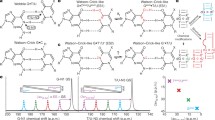
Here for the first time we present four novel routes of the tautomerisation via the sequential DPT that links biologically important A·C*(WC) DNA base mispair with Watson–Crick (WC) geometry and wobble (w) A*·C*(w), A·C*O2(w), A*·C*(w 1) and A·C(w ⊥) mismatches, pursuing the goal of estimation of their contribution into the transition mutations during DNA biosynthesis. These processes occur without opening of the pairs and are accompanied by the substantial changes in their geometry. A detailed analysis of these pathways leads to an identification of the A·C*(WC)↔A*·C*(w) tautomerisation route as the most suitable among these processes from the point of view of the spontaneous point mutagenesis, since it proceeds via the time that is significantly less than the time used by the replicative DNA-polymerase for the incorporation of one incoming nucleotide into the synthesised DNA double helix. This non-dissociative transition occurs through the planar, highly stable, zwitterionic \( {\text{TS}}_{{{\text{A}} \cdot {\text{C*(WC)}} \leftrightarrow {\text{A}}^{ + } \cdot {\text{C}}^{\text{ - }} (w )}}^{{{\text{A}}^{ + } \cdot {\text{C}}^{\text{ - }} }} \) transition state and dynamically unstable intermediate A+·C−(w) ion pair and is accompanied by the consistent rearrangement of the 10 unique patterns of the specific intermolecular interactions, among which there are from 2 to 4 AH···B H-bonds and 2 loosened A–H–B covalent bridges. Basic physico-chemical properties of this mutual tautomeric transformation, which is internally inherent to the A·C*(WC) and A*·C*(w) base mispairs, are documented, and its possible biological assignment is discussed here.









Similar content being viewed by others
References
Jacquemin D, Zúñiga J, Requena A, Céron-Carrasco JP (2014) Assessing the importance of proton transfer reactions in DNA. Acc Chem Res 47:2467–2474
Brovarets’ OO, Hovorun DM (2013) Atomistic understanding of the C·T mismatched DNA base pair tautomerization via the DPT: QM and QTAIM computational approaches. J Comput Chem 34:2577–2590
Brovarets’ OO, Zhurakivsky RO, Hovorun DM (2014) Is the DPT tautomerisation of the long A·G Watson–Crick DNA base mispair a source of the adenine and guanine mutagenic tautomers? A QM and QTAIM response to the biologically important question. J Comput Chem 35:451–466
Watson JD, Crick FHC (1953) The structure of DNA. Cold Spring Harbor Symp Quant Biol 18:123–131
Topal MD, Fresco JR (1976) Complementary base pairing and the origin of substitution mutations. Nature 263:285–289
Brovarets’ OO, Kolomiets’ IM, Hovorun DM (2012) Elementary molecular mechanisms of the spontaneous point mutations in DNA: a novel quantum-chemical insight into the classical understanding. In: Tada T (ed) Quantum chemistry: molecules for innovations. In Tech Open Access, Rijeka
Cerón-Carrasco JP, Zúñiga J, Requena A, Perpète EA, Michaux C, Jacquemin D (2011) Combined effect of stacking and solvation on the spontaneous mutation in DNA. Phys Chem Chem Phys 13:14584–14589
Ceron-Carrasco JP, Jacquemin D (2015) DNA spontaneous mutation and its role in the evolution of GC-content: assessing the impact of the genetic sequence. Phys Chem Chem Phys 17:7754–7760
Brovarets’ OO, Hovorun DM (2014) Can tautomerisation of the A· T Watson–Crick base pair via double proton transfer provoke point mutations during DNA replication? A comprehensive QM and QTAIM analysis. J Biomol Struct Dyn 32:127–154
Brovarets’ OO, Hovorun DM (2014) Why the tautomerization of the G·C Watson–Crick base pair via the DPT does not cause point mutations during DNA replication? QM and QTAIM comprehensive analysis. J Biomol Struct Dyn 32:1474–1499
Danilov VI, Anisimov VM, Kurita N, Hovorun D (2005) MP2 and DFT studies of the DNA rare base pairs: the molecular mechanism of the spontaneous substitution mutations conditioned by tautomerism of bases. Chem Phys Lett 412:285–293
Fonseca Guerra C, Bickelhaupt FM, Saha S, Wang F (2006) Adenine tautomers: relative stabilities, ionization energies, and mismatch with cytosine. J Phys Chem A 110:4012–4020
Wang W, Hellinga HW, Beese LS (2011) Structural evidence for the rare tautomer hypothesis of spontaneous mutagenesis. Proc Natl Acad Sci USA 124:17644–17648
Brovarets’ OO, Hovorun DM (2015) The physicochemical essence of the purine·pyrimidine transition mismatches with Watson–Crick geometry in DNA: A·C* versa A*·C. A QM and QTAIM atomistic understanding. J Biomol Struct Dyn 33:28–55
Brovarets’ OO, Hovorun DM (2010) Quantum chemical investigation of the elementary molecular mechanisms of pyrimidine·purine transversions. Ukrains’kyi Biokhimichnyi Zhurnal 82:55–60
Brovarets’ OO, Hovorun DM (2009) Physicochemical mechanism of the wobble DNA base pairs Gua·Thy and Ade·Cyt transition into the mismatched base pairs Gua*·Thy and Ade·Cyt* formed by the mutagenic tautomers. Ukr Bioorg Acta 8:12–18
Brovarets’ OO, Hovorun DM (2010) How stable are the mutagenic tautomers of DNA bases? Biopolym Cell 26:72–76
Richard JP, Amyes TL (2004) On the importance of being zwitterionic: enzymatic catalysis of decarboxylation and deprotonation of cationic carbon. Bioorganic Chem 32:354–366
Brovarets’ OO, Hovorun DM (2015) Tautomeric transition between wobble A·C DNA base mispair and Watson–Crick-like A·C* mismatch: microstructural mechanism and biological significance. Phys Chem Chem Phys 17:15103–15110
Yang Y, Qin Y, Ding Q, Bakhtina M, Wang L, Tsai M-D, Zhong D (2014) Ultrafast water dynamics at the interface of the polymerase—DNA binding complex. Biochemistry 53:5405–5413
Brovarets’ OO, Yurenko YP, Dubey IY, Hovorun DM (2012) Can DNA-binding proteins of replisome tautomerize nucleotide bases? Ab initio model study. J Biomol Struct Dyn 29:1101–1109
Brovarets OO, Hovorun DM (2014) Does the G·G*syn DNA mismatch containing canonical and rare tautomers of the guanine tautomerise through the DPT? A QM/QTAIM microstructural study. Mol Phys 112:3033–3046
Rothwell PJ, Waksman G (2005) Structure and mechanism of DNA polymerases. Adv Protein Chem 71:401–440
Brovarets’ OO, Hovorun DM (2015) How many tautomerisation pathways connect Watson–Crick-like G*·T DNA base mispair and wobble mismatches? J Biomol Struct Dyn 33:2297–2315
Kirmizialtin S, Nguyen V, Johnson KA, Elber R (2012) How conformational dynamics of DNA polymerase select correct substrates: experiments and simulations. Structure 20:618–627
Cerón-Carrasco JP, Cerezo J, Jacquemin D (2014) How DNA is damaged by external electric fields: selective mutation versus random degradation. Phys Chem Chem Phys 16:8243–8246
Cerón-Carrasco JP, Jacquemin D (2013) Electric field induced DNA damage: an open door for selective mutations. Chem Commun 49:7578–7580
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Pople JA (2010) GAUSSIAN 09 (Revision B.01). Gaussian Inc, Wallingford
Tirado-Rives J, Jorgensen WL (2008) Performance of B3LYP density functional methods for a large set of organic molecules. J Chem Theory Comput 4:297–306
Parr RG, Yang W (1989) Density-functional theory of atoms and molecules. Oxford University Press, Oxford
Lee C, Yang W, Parr RG (1988) Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter Mater Phys 37:785–789
Lozynski M, Rusinska-Roszak D, Mack H-G (1998) Hydrogen bonding and density functional calculations: the B3LYP approach as the shortest way to MP2 results. J Phys Chem A 102:2899–2903
Matta CF (2010) How dependent are molecular and atomic properties on the electronic structure method? Comparison of Hartree–Fock, DFT, and MP2 on a biologically relevant set of molecules. J Comput Chem 31:1297–1311
Brovarets’ OO, Hovorun DM (2010) Stability of mutagenic tautomers of uracil and its halogen derivatives: the results of quantum-mechanical investigation. Biopolym Cell 26:295–298
Samijlenko SP, Yurenko YP, Stepanyugin AV, Hovorun DM (2012) Tautomeric equilibrium of uracil and thymine in model protein–nucleic acid contacts. Spectroscopic and quantum chemical approach. J Phys Chem B 114:1454–1461
Furmanchuk A, Isayev O, Gorb L, Shishkin OV, Hovorun DM, Leszczynski J (2011) Novel view on the mechanism of water-assisted proton transfer in the DNA bases: bulk water hydration. Phys Chem Chem Phys 13:4311–4317
Brovarets’ OO, Hovorun DM (2014) How the long G·G* Watson–Crick DNA base mispair comprising keto and enol tautomers of the guanine tautomerises? The results of the QM/QTAIM investigation. Phys Chem Chem Phys 6:15886–15899
Brovarets’ OO, Zhurakivsky RO, Hovorun DM (2013) The physico-chemical mechanism of the tautomerisation via the DPT of the long Hyp*·Hyp Watson–Crick base pair containing rare tautomer: a QM and QTAIM detailed look. Chem Phys Lett 578:126–132
Palafox MA (2014) Molecular structure differences between the antiviral nucleoside analogue 5-iodo-2′-deoxyuridine and the natural nucleoside 2′-deoxythymidine using MP2 and DFT methods: conformational analysis, crystal simulations, DNA pairs and possible behavior. J Biomol Struct Dyn 32:831–851
El-Sayed AA, Tamara Molina A, Alvarez-Ros MC, Alcolea Palafox M (2015) Conformational analysis of the anti-HIV Nikavir prodrug: comparisons with AZT and thymidine, and establishment of structure-activity relationships/tendencies in other 6′-derivatives. J Biomol Struct Dyn 33:723–748
Peng C, Ayala PY, Schlegel HB, Frisch MJ (1996) Using redundant internal coordinates to optimize equilibrium geometries and transition states. J Comput Chem 17:49–56
Frisch MJ, Head-Gordon M, Pople JA (1990) Semi-direct algorithms for the MP2 energy and gradient. Chem Phys Lett 166:281–289
Hariharan PC, Pople JA (1973) The influence of polarization functions on molecular orbital hydrogenation energies. Theor Chim Acta 28:213–222
Krishnan R, Binkley JS, Seeger R, Pople JA (1980) Self-consistent molecular orbital methods. XX. A basis set for correlated wave functions. J Chem Phys 72:650–654
Kendall RA, Dunning TH Jr, Harrison RJ (1992) Electron affinities of the first-row atoms revisited. Systematic basis sets and wave functions. J Chem Phys 96:6796–6806
Hratchian HP, Schlegel HB (2005) Finding minima, transition states, and following reaction pathways on ab initio potential energy surfaces. In: Dykstra CE, Frenking G, Kim KS, Scuseria G (eds) Theory and applications of computational chemistry: the first 40 years. Elsevier, Amsterdam, pp 195–249
Brovarets’ OO, Zhurakivsky RO, Hovorun DM (2013) The physico-chemical “anatomy” of the tautomerisation through the DPT of the biologically important pairs of hypoxanthine with DNA bases: QM and QTAIM perspectives. J Mol Model 19:4119–4137
Brovarets’ OO, Zhurakivsky RO, Hovorun DM (2014) Does the tautomeric status of the adenine bases change under the dissociation of the A*·Asyn Topal-Fresco DNA mismatch? A combined QM and QTAIM atomistic insight. Phys Chem Chem Phys 16:3715–3725
Boys SF, Bernardi F (1970) The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol Phys 19:553–566
Gutowski M, Van Lenthe JH, Verbeek J, Van Duijneveldt FB, Chalasinski G (1986) The basis set superposition error in correlated electronic structure calculations. Chem Phys Lett 124:370–375
Sordo JA, Chin S, Sordo TL (1988) On the counterpoise correction for the basis set superposition error in large systems. Theor Chim Acta 74:101–110
Sordo JA (2001) On the use of the Boys–Bernardi function counterpoise procedure to correct barrier heights for basis set superposition error. J Mol Struct THEOCHEM 537:245–251
Atkins PW (1998) Physical chemistry. Oxford University Press, Oxford
Brovarets’ OO, Zhurakivsky RO, Hovorun DM (2010) Is there adequate ionization mechanism of the spontaneous transitions? Quantum-chemical investigation. Biopolym Cell 26:398–405
Kosenkov D, Kholod Y, Gorb L, Shishkin O, Hovorun DM, Mons M, Leszczynski J (2009) Ab initio kinetic simulation of gas-phase experiments: tautomerization of cytosine and guanine. J Phys Chem B 113:6140–6150
Wigner E (1932) Über das Überschreiten von Potentialschwellen bei chemischen Reaktionen [Crossing of potential thresholds in chemical reactions]. Z Phys Chem B19:203–216
Brovarets’ OO, Hovorun DM (2013) Atomistic nature of the DPT tautomerisation of the biologically important C·C* DNA base mispair containing amino and imino tautomers of the cytosine: a QM and QTAIM approach. Phys Chem Chem Phys 15:20091–20104
Brovarets’ OO, Zhurakivsky RO, Hovorun DM (2013) DPT tautomerization of the long A·A* Watson–Crick base pair formed by the amino and imino tautomers of adenine: combined QM and QTAIM investigation. J Mol Model 19:4223–4237
Bader RFW (1990) Atoms in molecules: a quantum theory. Oxford University Press, Oxford
Keith TA (2010) AIMAll (Version 10.07.01). Retrieved from aim.tkgristmill.com
Brovarets’ OO, Yurenko YP, Hovorun DM (2014) Intermolecular CH···O/N H-bonds in the biologically important pairs of natural nucleobases: a thorough quantum-chemical study. J Biomol Struct Dyn 32:993–1022
Brovarets’ OO, Yurenko YP, Hovorun DM (2014) The significant role of the intermolecular CH···O/N hydrogen bonds in governing the biologically important pairs of the DNA and RNA modified bases: a comprehensive theoretical investigation. J Biomol Struct Dyn 33:1624–1652
Espinosa E, Molins E, Lecomte C (1998) Hydrogen bond strengths revealed by topological analyses of experimentally observed electron densities. Chem Phys Lett 285:170–173
Mata I, Alkorta I, Espinosa E, Molins E (2011) Relationships between interaction energy, intermolecular distance and electron density properties in hydrogen bonded complexes under external electric fields. Chem Phys Lett 507:185–189
Iogansen AV (1999) Direct proportionality of the hydrogen bonding energy and the intensification of the stretching ν(XH) vibration in infrared spectra. Spectrochim Acta Part A Mol Biomol Spectrosc 55:1585–1612
Nikolaienko TY, Bulavin LA, Hovorun DM (2012) Bridging QTAIM with vibrational spectroscopy: the energy of intramolecular hydrogen bonds in DNA-related biomolecules. Phys Chem Chem Phys 14:7441–7447
Saenger W (1984) Principles of nucleic acid structure. Springer, New York
Moore WJ (1983) Basic physical chemistry. Prentice-Hall, Englewood Cliffs
Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T (2006) DNA repair and mutagenesis. ASM Press, Washington
Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P (2002) Chapter 5. The initiation and completion of DNA replication in chromosomes. In: Molecular biology of the cell (4th edition), ISBN-10:0-8153-3218-1I, SBN-10:0-8153-4072-9, Garland Science, New York
Huang M-M, Arnheim N, Goodman M (1992) Extension of base mispairs by Taq DNA polymerase: implications for single nucleotide discrimination in PCR. Nucleic Acids Res 20:4567–4573
Fijalkowska IJ, Schaaper RM, Jonczyk P (2012) DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair. FEMS Microbiol Rev 36:1105–1121
Alemán EA, de Silva C, Patrick EM, Musier-Forsyth K, Rueda D (2014) Single-molecule fluorescence using nucleotide analogs: a proof-of-principle. J Phys Chem Lett 5:777–781
Kornienko IV, Malyarchuk BA (2005) Analysis of mutation mechanisms in human mitochondrial DNA. Mol Biol 39:761–768
Govorun DN, Danchuk VD, Mishchuk YR, Kondratyuk IV, Radomsky NF, Zheltovsky NV (1992) AM1 calculation of the nucleic acid bases structure and vibrational spectra. J Mol Struct 267:99–103
Pelmenschikov A, Hovorun DM, Shishkin OV, Leszczynski J (2000) A density functional theory study of vibrational coupling between ribose and base rings of nucleic acids with ribosyl guanosine as a model system. J Chem Phys 113:5986–5990
Nikolaienko TY, Bulavin LA, Hovorun DM (2011) How flexible are DNA constituents? The quantum-mechanical study. J Biomol Struct Dyn 29:563–575
Raczyńska ED, Makowski M, Zientara-Rytter K, Kolczyńska K, Stępniewski TM, Hallmann M (2013) Quantum-chemical studies on the favored and rare tautomers of neutral and redox adenine. J Phys Chem 117:1548–1559
Raczyńska ED, Makowski M, Hallmann M, Kamińska B (2015) Geometric and energetic consequences of prototropy for adenine and its structural models: a review. RSC Adv 5:36587–36604
Giulia Rossetti G, Dans PD, Gomez-Pinto I, Ivani I, Gonzalez G, Orozco M (2015) The structural impact of DNA mismatches. Nucleic Acids Res 43:4309–4321
Acknowledgments
O.O.B. was supported by the Grant of the President of Ukraine to support scientific research of young scientists for 2015 year from the State Fund for Fundamental Research of Ukraine (Project No. GP/F61/028), by the Grant of the NAS of Ukraine for young scientists for the years 2015–2016 and by the Scholarship of the President of Ukraine for young scientists for the years 2014–2016 given to O.O.B. This work was performed using computational facilities of joint computer cluster of SSI “Institute for Single Crystals” of the National Academy of Sciences of Ukraine and Institute for Scintillation Materials of the National Academy of Sciences of Ukraine incorporated into Ukrainian National Grid. The authors sincerely thank Dr. Ivan S. Voiteshenko (Institute of High Technologies, Taras Shevchenko National University of Kyiv) and Dr. Fernando R. Clemente (Gaussian, Inc.) for their technical support of the work.
Author information
Authors and Affiliations
Corresponding author
Additional information
This paper is dedicated to outstanding Ukrainian Scientist Dr. Sci. Oleg Shishkin, who made a great contribution into the development of quantum chemistry in Ukraine and left a prominent scientific heritage.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Brovarets’, O.O., Hovorun, D.M. By how many tautomerisation routes the Watson–Crick-like A·C* DNA base mispair is linked with the wobble mismatches? A QM/QTAIM vision from a biological point of view. Struct Chem 27, 119–131 (2016). https://doi.org/10.1007/s11224-015-0687-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-015-0687-4