Abstract
We consider the problem of surrogate sufficient dimension reduction, that is, estimating the central subspace of a regression model, when the covariates are contaminated by measurement error. When no measurement error is present, a likelihood-based dimension reduction method that relies on maximizing the likelihood of a Gaussian inverse regression model on the Grassmann manifold is well-known to have superior performance to traditional inverse moment methods. We propose two likelihood-based estimators for the central subspace in measurement error settings, which make different adjustments to the observed surrogates. Both estimators are computed based on maximizing objective functions on the Grassmann manifold and are shown to consistently recover the true central subspace. When the central subspace is assumed to depend on only a few covariates, we further propose to augment the likelihood function with a penalty term that induces sparsity on the Grassmann manifold to obtain sparse estimators. The resulting objective function has a closed-form Riemann gradient which facilitates efficient computation of the penalized estimator. We leverage the state-of-the-art trust region algorithm on the Grassmann manifold to compute the proposed estimators efficiently. Simulation studies and a data application demonstrate the proposed likelihood-based estimators perform better than inverse moment-based estimators in terms of both estimation and variable selection accuracy.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
1 Introduction
In a regression setting with an outcome \(y \in \mathbb {R}\) and a covariate vector \(\textbf{X}\in \mathbb {R}^p\), sufficient dimension reduction (SDR) refers to a class of methods that express the outcome as a few linear combinations of \(\textbf{X}\) (Li 2018). In other words, SDR aims to estimate the matrix \(\textbf{B}\in \mathbb {R}^{p \times d}\) such that \(y \perp \textbf{X}\mid \textbf{B}^{\text {T}} \textbf{X}\), where \(d \ll p \). As the matrix \(\textbf{B}\) is generally not unique, the estimation target in SDR is the central subspace \(\mathcal {S}_{y \vert \textbf{X}}\), defined as the intersection of all the subspaces spanned by the columns of \(\textbf{B}\) satisfying the above conditional independence condition. This central subspace is unique under mild conditions (Glaws et al. 2020), and is characterized by the projection matrix associated with \(\textbf{B}\).
This article focuses on the problem of estimating the central subspace \(\mathcal {S}_{y \vert \textbf{X}}\) when the covariates \(\textbf{X}\) are measured with error. This problem is known in the literature as surrogate sufficient dimension reduction (Li and Yin 2007). Instead of observing a sample for \(\textbf{X}\), we observe a sample of \(\textbf{W}\), which are surrogates related to \(\textbf{X}\) by the classical additive measurement error model \(\textbf{W}= \textbf{X}+ \textbf{U}\), where \(\textbf{U}\) is a vector of measurement errors independent of \(\textbf{X}\) and which follows a p-variate Gaussian distribution with mean zero and covariance matrix \(\varvec{\Sigma }_u\). While more general measurement error models have also been proposed in the literature e.g., \(\textbf{W}= \textbf{A}\textbf{X}+ \textbf{U}\) for \(\textbf{A} \in \mathbb {R}^{r \times p}\) with \(r \ge p\) (Carroll and Li 1992; Li and Yin 2007; Zhang et al. 2014), we highlight that the classical measurement error model with \(\textbf{A} = \textbf{I}_p\) is the most widely used in practice (see Grace et al. 2021; Chen and Yi 2022; Chen 2023; Nghiem et al. 2023, for some recent examples) and so we focus on this case throughout the paper. Also, throughout the article we assume \(\varvec{\Sigma }_u\) is known; in practice, this covariance matrix may be estimated from auxiliary data, such as replicate observations, before subsequent analyses are conducted (Carroll et al. 2006). Our observed data thus consists of n pairs \((y_i, \textbf{W}_i^{\text {T}})\) with \(\textbf{W}_i = \textbf{X}_i + \textbf{U}_i\), \(i=1,\ldots , n\).
When the true covariates \(\textbf{X}_i\) are observed, there exists a vast literature on how to estimate the central subspace; see Li (2018) for an overview. These include traditional inverse moment-based methods such as sliced inverse regression (Li 1991) and sliced average variance estimation (Cook 2000), forward regression methods such as minimum average variance estimation (Xia et al. 2002) and directional regression (Li and Wang 2007), and inverse regression methods such as principal fitted components (Cook and Forzani 2008) and likelihood-acquired directions (LAD, Cook and Forzani 2009). Recent literature has also expanded these methods into more complex settings, such as high-dimensional data (e.g., Lin et al. 2021; Qian et al. 2019) and longitudinal data (e.g., Hui and Nghiem 2022). Among the estimators discussed above, the LAD method developed by Cook and Forzani (2009) has a unique advantage in that it is constructed from a well-defined Gaussian likelihood function and, as such, inherits the optimality properties of likelihood theory. Also, while the conditional Gaussianity assumption appears unnatural, Cook and Forzani (2009) show that the LAD estimator has superior performance even when Gaussianity does not hold; roughly speaking, the conditional normality plays the role of a “working model.” In this article, we focus on adapting LAD to the surrogate SDR problem.
The problem of surrogate SDR was first considered in Carroll and Li (1992), who showed that \(\mathcal {S}_{y \vert \textbf{X}}\) can be estimated by performing ordinary least squares, or sliced inverse regression, of the response on the adjusted surrogate \(\textbf{X}^* = \textbf{R}\textbf{W}\), where \(\textbf{R}= \varvec{\Sigma }_{xw}\varvec{\Sigma }_{w}^{-1} = (\varvec{\Sigma }_{w}-\varvec{\Sigma }_u)\varvec{\Sigma }_{w}^{-1}\). Here, \(\varvec{\Sigma }_{xw} = \text {Cov}(\textbf{X},\textbf{W})\) denotes the covariance matrix of \(\textbf{X}\) and \(\textbf{W}\) and \(\varvec{\Sigma }_{w} = \text {Var}(\textbf{W})\) denotes the variance-covariance matrix of \(\textbf{W}\). When \(\varvec{\Sigma }_u\) is known, these adjusted surrogates can be computed by replacing \(\varvec{\Sigma }_w\) with an appropriate estimator from the sample. This underlying idea was expanded into a broader, invariance law by Li and Yin (2007), who prove that if both \(\textbf{X}\) and \(\textbf{U}\) follow multivariate Gaussian distributions, then \(\mathcal {S}_{y \vert {\textbf{X}^*}} = \mathcal {S}_{y \vert \textbf{X}} \). As a result, under the assumption of Gaussianity of both \(\textbf{X}\) and \(\textbf{U}\), any consistent SDR method applied to y and the adjusted surrogate \({\textbf{X}}^*\) is also consistent. If \(\textbf{X}\) is non-Gaussian, this relationship is maintained if \(\textbf{B}^{\text {T}} \textbf{X}\) is approximately Gaussian. For instance, this is achieved when \(p \rightarrow \infty \) and each column of \(\textbf{B}\) is dense i.e., the majority of elements in each column are non-zero. On the other hand, when the number of covariates is large, it is often assumed that the central subspace is sparse (i.e., it depends only on a few covariates Lin et al. 2019), and so this Gaussian approximation may not be realistic or relevant in practice. Elsewhere, Zhang et al. (2012) examines the surrogate SDR problem when both the response and the covariates are distorted by multiplicative measurement errors, while Chen and Yi (2022) proposed a SDR method for survival data when the response is censored and the covariates are measured with error at the same time. Neither of these addresses the sparsity of the central subspace when p is large, and more broadly there is little direct research on inverse regression methods for SDR under measurement error.
In this paper, we propose two likelihood-based estimators of the central subspace \(\mathcal {S}_{y\vert \textbf{X}}\) from the surrogate data \((y_i, \textbf{W}_i^{\text {T}})\). For the first estimator, we directly follow the approach of LAD and model the inverse predictor \(\textbf{X}\mid y\) as following a Gaussian distribution, where the existence of the central subspace imposes multiple constraints on the parameters and the covariates are subject to additive measurement errors. Measurement error is then incorporated into this model in a straightforward manner. We construct a likelihood-based function for the semi-orthogonal bases of the central subspace, and show that maximizing this function requires solving an optimization problem on the Grassmann manifold. For the second estimator, we apply the LAD approach on the adjusted surrogate \(\textbf{X}^*\). Although the two estimators make different adjustments to the adjusted surrogates, we show that these two estimators are asymptotically equivalent in terms of estimating the true central subspace. Furthermore, we propose a sparse estimator for the central subspace by augmenting the likelihood function with a penalty term that regularizes the elements of the corresponding projection matrix. The resulting objective function has a closed-form Riemann gradient, which facilitates efficient computation of the penalized estimator. Simulation studies and an application to a National Health and Nutrition Examination Survey from the United States demonstrate that the performance of the proposed likelihood-based estimators is superior to several common inverse moment-based estimators in terms of both estimating the central subspace and variable selection accuracy.
The rest of the paper is organized as follows: In Sect. 2, we briefly review the LAD estimator of the central subspace, which is built upon a Gaussian inverse regression model. Then we incorporate measurement errors and discuss the properties of the corresponding model. We propose maximum likelihood estimators of the central subspace from the surrogate data in Sect. 3. In Sect. 4, we propose a regularized estimator to achieve sparsity in the estimated central subspace. Section 5 presents simulation studies to demonstrate the performance of the proposed estimators, while Sect. 6 illustrates the methodology in a data application. Finally, Sect. 7 contains some concluding remarks.
2 A Gaussian inverse regression model with measurement errors
We first review the LAD method proposed by Cook and Forzani (2009), when the true covariate vector \(\textbf{X}\) is observed. LAD models the inverse predictor \(\textbf{X}\mid y\) as having a multivariate Gaussian distribution with mean \(\varvec{\mu }_y\) and covariance \(\varvec{\Delta }_y\) i.e., \(\textbf{X}\mid y \sim N(\varvec{\mu }_y, \varvec{\Delta }_y)\). Importantly, the existence of the central subspace \(\mathcal {S}_{y \vert \textbf{X}}\) imposes some constraints on the parameters \(\varvec{\mu }_y\) and \(\varvec{\Delta }_y\). Let \(\varvec{\mu }= \text {E}(\textbf{X})\), \(\varvec{\Delta }= \text {E}(\varvec{\Delta }_y) = \text {E}\left\{ \text {Var}(\textbf{X}\mid y) \right\} \), and let \(\varvec{\Psi }\in \mathbb {R}^{p \times d}\) be a semi-orthogonal basis matrix for \(\mathcal {S}_{y \vert \textbf{X}}\) and \(\varvec{\Psi }_0 \in \mathbb {R}^{p \times (p-d)} \) be a matrix, such that \((\varvec{\Psi }, \varvec{\Psi }_0) \in \mathbb {R}^{p \times p}\) is an orthogonal matrix. Then \(\mathcal {S}_{y \vert \textbf{X}}\) is a central subspace if and only if the two following conditions are satisfied:
where \(\varvec{v}_y \in \mathbb {R}^d\) denotes a deterministic function of y, \(\textbf{H}= (\varvec{\Psi }_0^{\text {T}} \varvec{\Delta }\varvec{\Psi })(\varvec{\Psi }^{\text {T}}\varvec{\Delta }\varvec{\Psi })^{-1}\) and \(\textbf{D}= (\varvec{\Psi }_0^{\text {T}} \varvec{\Delta }^{-1} \varvec{\Psi }_0)^{-1}\). While condition (i) allows the conditional distribution of \(\varvec{\Psi }^{\text {T}} \textbf{X}\mid y\) to depend on y, condition (ii) requires that the conditional distribution \(\varvec{\Psi }_0^{\text {T}} \textbf{X}| (\varvec{\Psi }^{\text {T}} \textbf{X}, y)\) does not depend on y. This aligns with the intuition that \(\varvec{\Psi }^{\text {T}} \textbf{X}\) is a sufficient predictor for y. Cook and Forzani (2009) also provide several equivalent characterisations of the two conditions in (1), for example, \(\varvec{\Psi }_0^{\text {T}} \varvec{\Delta }_y^{-1} = \varvec{\Psi }_0^{\text {T}} \varvec{\Delta }^{-1}\) and \(\varvec{\mu }_y - \varvec{\mu }= \varvec{\Delta }\varvec{\Psi }\varvec{v}_y\). The advantage of the conditions in Eq. (1) is that a log-likelihood function can be constructed and we can maximize it to consistently estimate all the parameters of the LAD model. As reviewed in Sect. 1, Cook and Forzani (2009) highlighted that Gaussianity of the inverse predictor \(\textbf{X}\mid y\) is not essential for LAD. Indeed, their simulation results show that the LAD estimator has superior performance to other SDR methods in terms of recovering the central subspace, even when the Gaussianity assumption is not satisfied.
When measurement errors that follow the classical additive measurement error model are present in the covariates so \(\textbf{X}\) is replaced by \(\textbf{W}\), condition (ii) in (1) is no longer satisfied. Specifically, we can straightforwardly show that the conditional mean of \(\varvec{\Psi }_0^{\text {T}} \textbf{W}\) given \(\varvec{\Psi }^{\text {T}} \textbf{W}\) and y is \( \text {E} ( \varvec{\Psi }_0^{\text {T}} \textbf{W}\vert \varvec{\Psi }^{\text {T}} \textbf{W}, y) = \varvec{\Psi }_0^{\text {T}} \varvec{\mu }+ \varvec{\Psi }_0^{\text {T}} \varvec{\Delta }\varvec{\Psi }\varvec{v}_y - (\textbf{H}\varvec{\Psi }^{\text {T}} \varvec{\Delta }_y \varvec{\Psi }+ \varvec{\Psi }_0^{\text {T}} \varvec{\Sigma }_u \varvec{\Psi })\left\{ \varvec{\Psi }^{\text {T}} (\varvec{\Delta }_y + \varvec{\Sigma }_u) \varvec{\Psi }\right\} ^{-1}\) \((\varvec{\Psi }^{\text {T}}\textbf{W}- \varvec{\Psi }^{\text {T}} \varvec{\Delta }\varvec{\Psi }\varvec{v}_y)\). This quantity generally depends on y, and so \(\varvec{\Psi }\) is no longer guaranteed to be a sufficient dimension reduction of the central subspace \(\mathcal {S}_{y | W}\). By a similar argument, condition (ii) is also not satisfied when \(\textbf{X}\) is replaced by \(\hat{\textbf{X}} = \textbf{R}\textbf{W}\), with \(\textbf{R}= \varvec{\Sigma }_x \varvec{\Sigma }_w^{-1}\). We remark that this result does not contradict the invariance law of Li and Yin (2007), since that was established under the assumption that the marginal distribution of \(\textbf{X}\) is Gaussian. Here, the Gaussian inverse regression model assumes only that the conditional distribution of \(\textbf{X}\mid y\) is Gaussian.
To overcome the challenges of inverse regression-based SDR with measurement error, we propose a new predictor of \(\textbf{X}\) from the observed surrogate \(\textbf{W}\) which satisfies similar properties to those in (1), so that we can apply inverse regression based SDR. To this end, let \(\textbf{V}= \textbf{L}\textbf{W}\) with \(\textbf{L}= \varvec{\Delta }(\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1}\). The the result below ensures that if \(\varvec{\Psi }\) is a semi-orthogonal basis matrix for \(\mathcal {S}_{y \vert \textbf{X}}\), then it is also a semi-orthogonal basis matrix for \(S_{y \vert \textbf{V}}\).
Proposition 1
Let \(\textbf{G}_y = \textbf{L}(\varvec{\Delta }_y + \varvec{\Sigma }_u) \textbf{L}^{\text {T}}\) and \(\tilde{\textbf{D}} = (\varvec{\Psi }_0^{\text {T}} \varvec{\Delta }^{-1} \textbf{L}^{-1} \varvec{\Psi }_0)^{-1}\). The conditional distributions \(\varvec{\Psi }^{\text {T}} \textbf{V}| y\) and \(\varvec{\Psi }_0^{\text {T}} \textbf{V}\vert (\varvec{\Psi }^{\text {T}} \textbf{V}, y)\) are given by
The proof of Proposition 1 and all the other theoretical results can be found in the Appendix. In summary, \(\textbf{V}\) is purposefully constructed such that the conditional mean of \(\varvec{\Psi }^{\text {T}} \textbf{V}\vert y\) depends on y, but the conditional mean of \(\varvec{\Psi }_0^{\text {T}} \textbf{V}\vert (\varvec{\Psi }^{\text {T}} \textbf{V}, y)\) does not.
3 Maximum likelihood estimation
3.1 Corrected LAD estimator
We can now consider the problem of estimating the central subspace \(\mathcal {S}_{y \vert \textbf{X}}\) from the data \((y_i, \textbf{W}_i^{\text {T}})\), \(i=1,\ldots , n\). Similar to LAD and other SDR methods based on inverse-moments, we first partition the data into M non-overlapping slices based on the outcome data \(y_i\), for \(i=1,\ldots , n\). If the outcome is categorical, each slice corresponds to one category. If the outcome is continuous, these slices are constructed by dividing its range into M non-overlapping intervals. Let \(S_m \subset \{1, \ldots , n\}\) denote the index set of observations and \(n_m\) be the number of observations in the mth slice, \(m=1,\ldots ,M\). Next, assume the true covariate data within each slice are mutually independent and identically distributed, such that \( \textbf{X}_i^{(m)} \mid y_{i}^{(m)} \sim N(\varvec{\mu }_{y}^{(m)}, \varvec{\Delta }_{y}^{(m)})\) where \( \textbf{W}_i^{(m)} = \textbf{X}_i^{(m)} + \textbf{U}_i^{(m)}, ~ \textbf{U}_i^{(m)} \sim N(\varvec{0}, \varvec{\Sigma }_u), \) and we use the superscript m to index the slice to which the ith observation belongs. Furthermore, we set \(\text {E}\left( \varvec{\mu }_y^{(m)}\right) = \text {E}\left( \textbf{X}_i^{(m)}\right) = \varvec{\mu }\), and \(\text {E}\left( \varvec{\Delta }_y^{(m)}\right) = \varvec{\Delta }\), that is, these expectations do not depend on the slice m.
From Proposition 1, let \(\textbf{V}_{i}^{(m)} = \textbf{L}\textbf{W}_i^{(m)}\). Then we will estimate the central subspace of \(\mathcal {S}_{y \vert \textbf{X}}\) by maximizing the joint log-likelihood of \(\varvec{\Psi }^{\text {T}} \textbf{V}_i^{(m)}\) and \(\varvec{\Psi }_0^{\text {T}} \textbf{V}_i^{(m)}\) for \(i=1,\ldots , n\) and \(m=1,\ldots ,M\). Theorem 1 below gives the explicit form of this log-likelihood function when the dimension d is known.
Theorem 1
Let \(\varvec{\Psi }\in \mathbb {R}^{p \times d}\) be a semi-orthogonal basis matrix for \(\mathcal {S}_{y \vert \textbf{X}}\). Then the profile log-likelihood function for \(\varvec{\Psi }\) and \(\varvec{\Delta }\) from the observed data is given by
where \(\textbf{L}= \varvec{\Delta }(\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1}\), \(\tilde{\varvec{\Sigma }}_{w}\) and \(\tilde{\varvec{\Delta }}_{wm}\) are the sample (marginal) covariance matrix of \(\textbf{W}\) and the sample covariance matrix of \(\textbf{W}\) within the mth slice, and \(\vert \textbf{A}\vert \) denotes the determinant of the square matrix \(\textbf{A}\).
The profile likelihood in Theorem 1 depends on both \(\varvec{\Psi }\) and \(\varvec{\Delta }\), so maximizing it is challenging. We propose to estimate \(\varvec{\Delta }\) first using a method-of-moments approach as follows: First, ignoring the measurement errors, compute the naïve LAD estimator \(\hat{\mathcal {S}}_{n}\) on the observed data, \((y_i, {\textbf{W}}_i^{\text {T}})\), \(i=1,\ldots ,n\). Let \(\hat{\varvec{\Psi }}_{\text {n}}\) denote a subsequent orthogonal basis of \(\hat{\mathcal {S}}^{n}\). Then an estimate of \(\text {E}\left\{ \text {Var}(\textbf{W}\vert y) \right\} \) is given by \(\hat{\varvec{\Delta }}_{\text {n}} = \{\hat{\varvec{\Psi }}_{\text {n}}(\hat{\varvec{\Psi }}_{\text {n}}^{\text {T}} \tilde{\varvec{\Delta }}\varvec{\Psi }_{\text {n}})^{-1} \hat{\varvec{\Psi }}_{\text {n}}^{\text {T}} + \tilde{\varvec{\Sigma }}^{-1} - \hat{\varvec{\Psi }}_{\text {n}}(\hat{\varvec{\Psi }}_{\text {n}}^{\text {T}} \tilde{\varvec{\Sigma }}\varvec{\Psi }_{\text {n}})^{-1} \hat{\varvec{\Psi }}_{\text {n}}^{\text {T}} \}^{-1}\), and we can estimate \(\hat{\varvec{\Delta }}\) by \(\hat{\varvec{\Delta }}_{\text {n}} - \varvec{\Sigma }_u\). Replacing \(\varvec{\Delta }\) by \(\hat{\varvec{\Delta }}\) in (3) and removing terms that do not depend on \(\varvec{\Psi }\), we then maximize
where \( \hat{\textbf{L}} = \varvec{\Delta }(\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1}\). The right hand side of (4) is invariant to rotation of \(\varvec{\Psi }\) by an orthogonal matrix \(\textbf{A}\in \mathbb {R}^{d \times d}\). That is, for any matrix \(\textbf{A}\) with \(\textbf{A}^{\text {T}} = \textbf{A}^{-1}\), we have \(\ell _2(\varvec{\Psi }\textbf{A}) = \ell _2 (\varvec{\Psi })\). Therefore, the function in (4) is actually a function of the column space of \(\varvec{\Psi }\), which is characterized by the projection matrix \(P_{\mathcal {S}} = \varvec{\Psi }\varvec{\Psi }^{\text {T}}\). Similar to LAD then, maximisation of (4) is performed over the Grassmann manifold \( \mathcal {S} \in \mathcal {G}_{d, p} \subset \mathbb {R}^{p}\), which is the subspace spanned by any \(p \times d\) basis matrix. Specifically, the function in (4) can be written as
where for any square matrix \(\textbf{A}\), we use \(\Vert \textbf{A}\Vert _0\) to denote the product of its non-zero eigenvalues. The corresponding Euclidean gradient of this objective function at a semi-orthogonal basis \(\varvec{\Psi }\) of \(\mathcal {S}\) is
and the corresponding Riemann gradient of \(\ell _2(\mathcal {S})\) is given by \({\text {grad}}\{{\ell }_2(\mathcal {S})\} = (\textbf{I}_p - P_{\mathcal {S}}) \nabla {\ell }_2(\varvec{\Psi })\). Note the Euclidean gradient \(\nabla \ell _2(\varvec{\Psi })\) is derived by treating \(\varvec{\Psi }\) as a collection of (pd) unstructured elements, and the Riemann gradient is the projection of this Euclidean gradient onto the tangent space of \(\mathcal {G}_{d, p}\) at the subspace \(\mathcal {S}\) spanned by \(\varvec{\Psi }\).
This closed-form Riemann gradient function facilitates the use of a trust region algorithm on the Grassmann manifold developed by Absil et al. (2007). Starting from an initial solution \(\hat{\mathcal {S}}^{(0)}\), the algorithm finds the next candidate \(\hat{\mathcal {S}}^{(1)}\) by first finding a solution to minimize a constrained objective function on the tangent space at \(\hat{\mathcal {S}}^{(0)}\), and then maps this solution to \(\mathcal {G}_{d, p}\). The constraints imposed on the tangent space are known as a trust region. The decision to accept the candidate and to expand the region or not is based on a quotient; different values of the quotient lead to one out of three possibilities: (i) accept the candidate and expand the trust region, (ii) accept the candidate and reduce the trust region, or (iii) reject the candidate and reduce the trust region. This procedure is carried out until convergence; see Gallivan et al. (2003) and Absil et al. (2007) for further details. We use the Manopt package (Boumal et al. 2014) in Matlab, which is a dedicated toolbox for optimization on manifolds and matrices, to maximize (5). In this implementation, there is no need to provide the Hessian of \(\ell _2(\mathcal {S})\), since the Manopt package automatically incorporates a numerical approximation for this matrix based on finite differences in the trustregion solver.
For the remainder of this article, we refer to the solution of this problem, and hence our proposed estimator, as the corrected LAD estimator (cLAD) of the central subspace.
3.2 Invariance-law LAD estimator
The cLAD estimator is constructed by maximizing a likelihood function involving the adjusted surrogate \(\textbf{V}_i = \textbf{L}\textbf{W}_i = {\varvec{\Delta }} ({\varvec{\Delta }} + {\varvec{\Sigma }}_u)^{-1} \textbf{W}_i\). In this section, we consider a likelihood-based estimator for the central subspace based instead on maximizing a likelihood function involving \(\textbf{X}^* = {\varvec{\Sigma }}_x{\varvec{\Sigma }}_w^{-1}\textbf{W}\), the adjusted covariate introduced in the invariance law of Li and Yin (2007). From the observed data, a sample of the adjusted covariate \(\textbf{X}^*\) can be constructed by \(\widehat{\textbf{X}}_i^* = \hat{\varvec{\Sigma }}_x \hat{{\varvec{\Sigma }}}_w^{-1}\textbf{W}_i^{\top }\), where \(\hat{{\varvec{\Sigma }}}_{w} = n^{-1}\sum _{i=1}\textbf{W}_i \textbf{W}_i^{\text {T}}\) and \(\hat{{\varvec{\Sigma }}}_x = \hat{{\varvec{\Sigma }}}_{w} -\varvec{\Sigma }_u\). The equivalence between \(\mathcal {S}_{y\vert X}\) and \(\mathcal {S}_{y\vert X^*}\) motivates applying LAD to (\(y_i, \hat{\textbf{X}}_i^*\)), to obtain the invariance-law LAD (IL-LAD) estimator \(\hat{\varvec{\Psi }}^*\) which maximizes the objective function
where \(\tilde{\varvec{\Sigma }}^*\) and \(\tilde{\varvec{\Delta }}_m^*\) denote the sample marginal covariance of \(\textbf{X}^*\) and the sample covariance of \(\textbf{X}^*\) within the mth slice, respectively.
Similar to the cLAD estimator, maximization of 3.2 is also performed over the Grassmann manifold \( \mathcal {S} \in \mathcal {G}_{d, p}\). The main difference between the estimators is that the cLAD estimator uses the conditional covariance \({\varvec{\Delta }} = \text {E}\left\{ \text {Var}(\textbf{X}\mid y\right\} \) and the IL-LAD estimator uses the marginal covariance \({\varvec{\Sigma }}_x\) to construct the adjusted covariate. Nevertheless, in the following subsection, we will prove that the IL-LAD and cLAD estimator are asymptotically equivalent. In finite samples, we found that the difference between the two estimators is often negligible.
3.3 Consistency
In this section, we establish the consistency of both the LAD and IL-LAD estimators, and show them to be asymptotically equivalent. We focus on the setting when d is known and p is fixed, similar to Cook and Forzani (2009). Also, note that while we assume the measurement error covariance matrix \({\varvec{\Sigma }}_u\) to be known, we highlight that the theoretical results in this section will continue to hold if \({\varvec{\Sigma }}_u\) is replaced by a consistent estimator \(\hat{{\varvec{\Sigma }}}_u\). An example of the latter is given in our data example in Sect. 6, where replications are available for the n observations. When the true covariates \(\textbf{X}\) are observed, Cook and Forzani (2009) proved the consistency of the LAD estimator by establishing the equivalence between the population subspace spanned by LAD and that spanned by the sliced average variance estimator (SAVE), \(\mathcal {S}_{\text {SAVE}} = {\varvec{\Sigma }}_x^{-1} \text {span} \left( {\varvec{\Sigma }}_x - {\varvec{\Delta }}_1, \ldots , {\varvec{\Sigma }}_x - {\varvec{\Delta }}_m \right) \). We use a similar argument here and prove that, when \(n \rightarrow \infty \), the subspace spanned by either the cLAD or IL-LAD estimators is equivalent to that of the SAVE estimator when the true covariates X are observed.
For any subspace \(\mathcal {S}\), the function \(n^{-1}\ell _2(\mathcal {S})\) from (5) converges in probability to \( K_2(\mathcal {S}) = \log \Vert P_{\mathcal {S}} (\textbf{L}(\varvec{\Sigma }_x + \varvec{\Sigma }_u) \textbf{L}^{\text {T}}) P_{\mathcal {S}} \Vert _0 - \sum _{m=1}^{M} f_m \log \Vert P_{\mathcal {S}} {\textbf{L}} (\varvec{\Delta }_y^{(m)} + \varvec{\Sigma }_u) \textbf{L}^{\text {T}} P_{\mathcal {S}} \Vert _0\), where \( f_m = n_m/n. \) The population cLAD subspace is then defined to be \(\mathcal {S}^*_{\text {LAD}} = \arg \max _{S} K_2(\mathcal {S})\). Similarly, the subspace spanned by the IL-LAD estimator converges to the population IL-LAD subspace \(\mathcal {S}^*_{\text {IL-LAD}}\) that maximizes \( K_3(\mathcal {S}) = \log \Vert P_{\mathcal {S}} \varvec{\Sigma }^* P_{\mathcal {S}} \Vert _0 - \sum _{m=1}^{M} f_m \log \Vert P_{\mathcal {S}} \varvec{\Delta }_m^* P_{\mathcal {S}} \Vert _0,\) with \({\varvec{\Sigma }}^* = \text {Var}(\textbf{X}^*) = {\varvec{\Sigma }}_x{\varvec{\Sigma }}_w^{-1}{\varvec{\Sigma }}_x\), and \({\varvec{\Delta }}_m^* = \text {Var}\left( \textbf{X}_i^{(m)*} \mid y_i^{(m)} \right) = {\varvec{\Sigma }}_x {\varvec{\Sigma }}_{w}^{-1}\left( {\varvec{\Delta }}_y^{(m)}+ {\varvec{\Sigma }}_u\right) {\varvec{\Sigma }}_{w}^{-1}{\varvec{\Sigma }}_x\). The main result below establishes the equivalence between the population cLAD and IL-LAD subspaces and the population SAVE subspace.
Theorem 2
\(\mathcal {S}^*_{\textrm{cLAD}} = \mathcal {S}^*_{\mathrm {IL-LAD}} = \mathcal {S}_{\textrm{SAVE}}\).
As noted in Sect. 3.2, the main difference between the cLAD and the IL-LAD estimators is the use of \({\varvec{\Delta }}\) versus \({\varvec{\Sigma }}_x\) in the construction of the adjusted surrogate. Nevertheless, asymptotically, they are both equivalent to the true SAVE estimator, since the orthogonal complement \({\varvec{\Phi }}_0\) corresponding to the SAVE estimator satisfies \({\varvec{\Phi }}_0^{\text {T}}op {\varvec{\Delta }}^{-1} = {\varvec{\Phi }}_0^{\text {T}}op {\varvec{\Sigma }}_x^{-1}\). Similar to Proposition 2 in Cook and Forzani (2009), Theorem 2 does not require any distributional assumptions on the model, and only depends on the properties of positive definite matrices and the concavity of the log determinant function. Nevertheless, the result implies that the cLAD estimator is consistent whenever the true LAD estimator and SAVE (i.e., those computed assuming \(\textbf{X}\) were known) are consistent for the central subspace \(\mathcal {S}_{y \vert \textbf{X}}\). As shown in Li and Wang (2007) and Cook and Forzani (2009), these two estimators are consistent under a linearity and constant covariance condition.
4 Sparse surrogate dimension reduction
When the number of covariates is large, it is typically assumed that the sufficient predictors \(\textbf{B}^{\text {T}} \textbf{X}\) depend on only a few covariates (e.g., Qian et al. 2019). In other words, the true matrix \(\textbf{B}\) is row-sparse. Since only the column space of \(\textbf{B}\) is identifiable from the samples, we translate the sparsity of \(\textbf{B}\) into the element-wise sparsity of the projection matrix, and impose a corresponding penalty to achieve sparse solutions. Below, we formalize this idea with the proposed cLAD estimator, although an analogous procedure can be applied to the IL-LAD estimator.
We propose to maximize the regularized objective function
where for any matrix \(\textbf{A}\) with elements \(a_{ij}\), we denote \(\Vert \textbf{A}\Vert _1 = \sum _{i,j} \vert a_{ij} \vert \), and \(\lambda > 0\) is a tuning parameter. Although it is possible to use other regularization functions to achieve sparse solutions, here we follow Wang et al. (2017) and choose the \(\ell _1\) norm regularizer to ease the optimization problem on the Grassmann manifold, as both the Euclidean and Riemann gradient of the objective function \(\tilde{\ell }_2(\mathcal {S})\) have a closed form.
In more detail, for any matrix \(\textbf{A}\) of arbitrary dimension \(m \times n\), let \(\text {vec}(\textbf{A})\) denote the mn-dimensional vector formed by stacking columns of \(\textbf{A}\) together. Similarly, let \(\text {ivec}(\cdot )\) denote the inverse vectorization operator i.e., \({\text {ivec}}\{({\text {vec}}(\textbf{A})\} = \textbf{A}\). Let \(\textbf{T}_{m,n} \) be a (unique) matrix of dimension \(mn \times mn\) satisfying \(\text {vec}(\textbf{A}) = \textbf{T}_{m, n} \text {vec}(\textbf{A}^{\text {T}})\). Taking the Euclidean gradient of the regularization term with respect to \(\varvec{\Psi }\), we obtain
where \(\textbf{I}_{p^2}\) is the identity matrix of dimension \(p^2 \times p^2\) and \(\otimes \) denotes the Kronecker product. As a result, the Euclidean gradient of the \(\tilde{\ell }_2(\mathcal {S})\) at any semi-orthogonal matrix \(\varvec{\Psi }\) is \( \nabla \tilde{\ell }_2(\varvec{\Psi }) = \nabla \ell _2(\varvec{\Psi }) - \lambda \,\text {ivec} \left\{ \text {vec}\left( \left\| \varvec{\Psi }\varvec{\Psi }^{T}\right\| _{1}/\partial \varvec{\Psi }\right) \right\} , \) and the corresponding Riemann gradient is \( {\text {grad}}\tilde{\ell }_2(\mathcal {S}) = (\textbf{I}_p - \varvec{\Psi }\varvec{\Psi }^{\text {T}}) \nabla \tilde{\ell }_2(\varvec{\Psi }). \) These Euclidean and Riemann gradients are used in the same trust region algorithm of Absil et al. (2007) to maximize \(\tilde{\ell }_2(\mathcal {S})\). We computed the maximizer for (6) on a grid of the tuning parameter \(\lambda \) that consists of 40 logarithmically equally-spaced values between 0 and \(\lambda _{\text {max}}\), where \(\lambda _{\text {max}}\) is the value where the maximizer \(P_{\hat{\mathcal {S}}}\) of (6) is approximately close to the identity matrix. In our experiment, we found that setting \(\lambda _{\max }\) to 1 leads to a good performance and that the algorithm converges quickly to a stable solution at each value of the tuning parameter \(\lambda \) in the grid.
Finally, to select the optimal tuning parameter \(\lambda \), we use a variant of the projection information criterion (PIC) developed by Nghiem et al. (2022). Let \(\hat{\mathcal {S}}_0\) be an initial consistent, non-sparse estimator of the central subspace, and let \(\hat{\mathcal {S}}(\lambda )\) be the maximizer of (6) associated with \(\lambda \). We propose to select \(\lambda \) by minimizing \( \textsc {PIC}(\lambda ) = \Vert P_{\hat{\mathcal {S}}(\lambda )} - P_{\hat{S}_0}\Vert _F^2 + p^{-1}\log (p) {\text {df}}(P_{\hat{\mathcal {S}}(\lambda )}), \) where \({\text {df}}(P_{\hat{\mathcal {S}}(\lambda )}) = d(s_{\lambda }-d)\) with \(s_{\lambda }\) being the number of non-zero, diagonal elements of \(P_{\hat{\mathcal {S}}(\lambda )}\). The first term in \(\textsc {PIC}(\lambda )\) is a measure of goodness-of-fit, while the second term is a measure of complexity. Compared to the information criterion developed in Nghiem et al. (2022), this modified criterion has a different complexity term so as to more effectively quantify the number of parameters of the corresponding Grassmann manifold. In our numerical studies, we choose the initial consistent estimator \(\hat{\mathcal {S}}_0\) to be the estimated central subspace corresponding to the unregularized cLAD estimator, and find that this choice usually leads to good variable selection performance. For the remainder of this article, we refer to the maximizer of (6), with the tuning parameter selected via this projection criterion, as the sparse corrected LAD (scLAD) estimator.
5 Simulation studies
We conduct a numerical study to examine the performance of the proposed cLAD and IL-LAD estimators in finite samples. We generate the true predictors and outcome from the following four single/multi-index models (i) \(y_i = (0.5)(\textbf{X}_i^{\text {T}} \varvec{\beta }_1)^3 + 0.25~ \vert \textbf{X}_i^{\text {T}} \varvec{\beta }\vert \varepsilon _i\), (ii) \(y_i = 3(\textbf{X}_i^{\text {T}} \varvec{\beta }_1)/(1+\textbf{X}_i^{\text {T}} \varvec{\beta }_1)^2 + 0.25 \varepsilon _i\), (iii) \(y_i = 4\sin (\textbf{X}_i^{\text {T}} \varvec{\beta }_2/4) + 0.5(\textbf{X}_i^{\text {T}} \varvec{\beta }_1)^2 + 0.25\varepsilon _i\), and (iv) \(y_i = 3(\textbf{X}_i^{\text {T}} \varvec{\beta }_1)\exp (\textbf{X}_i^{\text {T}} \varvec{\beta }_2 + 0.25 \varepsilon _i) \), with \(\varepsilon _i \sim N(0,1)\), and \(\textbf{W}_i = \textbf{X}_i + \textbf{U}_i\) for \(i=1,\ldots , n\). The single index models (i) and (ii) are similar to those considered in Lin et al. (2019) and Nghiem et al. (2022), while the multiple index models (iii) and (iv) are similar to those considered in Reich et al. (2011). The true central subspace in models (i) and (ii) is the subspace spanned by \(\textbf{B}= \varvec{\beta }_1\), while in models (iii) and (iv) it is the column space of \(\textbf{B}= [\varvec{\beta }_1, \varvec{\beta }_2]\). Moreover, we set \(\varvec{\beta }_1 = (1, 1, 1, 0, \ldots , 0)^{\text {T}}\) and \(\varvec{\beta }_2 = (0, 0, 1, 1, 1, 0, \ldots , 0)^{\text {T}}\) such that the true central subspace for the single index models depends only on the first three covariates, while for the multiple index models it depends only on the first five covariates across two indices.
Next, we generate the true predictors \(\textbf{X}_i\) from one of the following three choices: (1) a p-variate Gaussian distribution \(N(\varvec{0}, \varvec{\Sigma }_x)\), (2) a p-variate t distribution with three degrees of freedom and the same covariance matrix \(\varvec{\Sigma }_x\), and (3) a p-variate half-Gaussian distribution \(\vert N(\varvec{0}, \varvec{\Sigma }_x) \vert \), where for all three choices we set the covariance matrix to have an autoregressive structure \(\sigma _{xij} = 0.5^{\vert i-j \vert }\). Turning to the measurement error, we generate \(\textbf{U}_i\) from a multivariate Gaussian distribution \(N(\varvec{0}, \varvec{\Sigma }_u)\), where \(\varvec{\Sigma }_u\) is set to a diagonal matrix with elements drawn from a uniform U(0.2, 0.5) distribution. The sample size n is set to either 1000 or 2000, while the number of covariates p is set to either 20 or 40. We assume that \(\varvec{\Sigma }_u\) and the structural dimension d are known. For each simulation configuration, we generated 200 samples.
We point out that the data generation process used for the simulation study is often referred to as a forward index model, because we first generate the covariate \(\textbf{X}\) from a marginal distribution, and then generate the response y from a conditional distribution i.e., from \(y\mid \textbf{X}\), which is the same as \(y \mid \textbf{B}^{\text {T}}op \textbf{X}\). More importantly, we highlight that the data generation processes are not the same as those imposed by the Gaussian inverse regression models. Particularly, with these simulation configurations, the conditional distribution \(\textbf{X}_i \mid y_i\) is generally not Gaussian due to the presence of non-linear link functions. As such, with these simulation configurations, the conditional Gaussianity only plays the role of a “working model.” This type of data generation process is also used in Cook and Forzani (2009).
For each simulated dataset, we compute the two unregularized likelihood-based estimates, cLAD and IL-LAD, and compare them with several invariance-law inverse-moment-based estimates, including SIR (IL-SIR), the SAVE (IL-SAVE) and directional regression (IL-DR). To account for the sparsity of the central subspace, we compute the scLAD estimator and compare it with two of the invariance-law sparse estimates, namely the Lasso SIR of Lin et al. (2019) (IL-Lin) and the sparse SIR proposed by Tan et al. (2018) (IL-Tan). Both of these estimators are sparse versions of sliced inverse regression, where the first imposes sparsity on each sufficient direction and the second imposes sparsity on the projection matrix and induces row-sparsity. For the estimator proposed by Lin et al. (2019), we use the R package LassoSIR with its default settings. For the estimator proposed by Tan et al. (2018), we use the code provided by the authors. For both estimators, we follow the recommend procedure from the authors and choose the tuning parameter based on a ten-fold cross-validation procedure.
We assess the performance of all estimators based on the Frobenius norm of the difference between the projection matrix associated with the true central subspace, and that of the estimated central subspace. For the sparse estimators, we assess variable selection performance based on the following metric. Let \(\textbf{P}\) denote the projection matrix associated with the true directions, i.e., \(\textbf{P} = \textbf{B}\left( \textbf{B}^{\text {T}} \textbf{B}\right) ^{-1}\textbf{B}^{\text {T}}\), and let \(\hat{\textbf{P}}\) be an estimator of \(\textbf{P}\). We then define the number of true positives (TP) to be the number of non-zero diagonal elements that are correctly identified to be non-zeros, the number of false negatives (FN) to be the number of non-zero diagonal elements that are incorrectly identified to be zeros and the number of false positives (FP) to be the number of zero diagonal elements that are incorrectly identified to be non-zeros. We also compute the F1 score as 2TP/(2TP + FP + FN). This metric ranges from zero to one, with a value of one indicating perfect variable selection.
For brevity, we present the projection error and F1 variable selection results for \(p=40\) below; the results for \(p=20\) and including FP and FN selection results offer overall similar conclusions to those seen below and are deferred to the Supplementary Materials. Table 1 demonstrates that among the unregularized estimators, the likelihood-based estimators cLAD and IL-LAD have smaller estimation error in the majority of settings compared to the other, invariance-law inverse-moment-based estimators. This result is consistent with Cook and Forzani (2009) who demonstrated that LAD, in general, tends to exhibit superior performance relative to other inverse-moment-based estimators like SIR and SAVE when no measurement error is present. The performance of all the considered estimators tends to deteriorate when the true covariates X deviate from Gaussianity, such as when they are skewed or have heavier tails. There is negligible difference in the performance between the cLAD and IL-LAD estimator.
Next, the left half of Table 2 demonstrates the estimation performance of the sparse SDR estimators. In terms of projection error, and analogous to the results with unregularized methods above, the proposed scLAD estimator tends to have lower estimation error than the IL-Lin and IL-Tan estimators. The improvement is most pronounced in the two multiple index models, i.e. Model (iii) and Model (iv), and when the true covariate-vector \(\textbf{X}\) follows a Gaussian distribution. Again similar to the unregularized estimators, the performance of these sparse estimators deteriorates when the true covariates \(\textbf{X}\) deviate from Gaussianity, especially when they follow a heavy tail distribution such as \(t_3\). As an aside, we note that results presented in Table 2 for the scLAD estimator represent the contributions of both the likelihood and sparsity to estimation performance. The results for the contribution of the likelihood function alone i.e., the unregularized cLAD estimator, were presented in Table , and for reasons of brevity we chose to avoid explicitly repeating these results in Table 2.
Turning to the variable selection results, the scLAD estimator had the best overall selection performance among the three considered estimators. When the true covariate vector \(\textbf{X}\) follows a Gaussian distribution, the scLAD estimator selects the true set of important covariates across all considered settings, as reflected in the corresponding F1 scores all being exactly equal to one.
This conclusion still holds when the true covariates follow a half-Gaussian distribution and under the single index models (i) and (ii). However, for the other settings such as the multiple index models (iii) and (iv), scLAD tends to have lower F1 scores than IL-Tan for \(n=1000\), although the trend is reversed when the sample size increases to \(n=2000\). By contrast, the IL-Lin estimator consistently has a very low F1 score, with additional results in the Supplementary Materials demonstrating that this estimator incurs too many false positives. This reflects the advantage of imposing regularisation directly on the diagonal elements of the corresponding projection matrix, compared to doing so on each dimension separately. However, we acknowledge that the variable selection results do depend on how the tuning parameter is chosen, and we are not aware of any method that is guaranteed to achieve selection consistency when we have to estimate the central subspace from surrogates.
6 National health and nutrition examination survey data
We apply the proposed methodology to analyze a dataset from the National Health and Nutrition Examination Survey (NHANES) (Centers for Disease Control and Prevention 2022). This survey aims to assess the health and nutritional status of people in the United States and track the evolution of this status over time. During the 2009-2010 survey period, participants were interviewed and asked to provide their demographic background as well as information about nutrition habits, and to undertake a series of health examinations. To assess the nutritional habits of participants, dietary data were collected using two 24-hour recall interviews wherein the participants self-reported the consumed amounts of a set of food items during the 24 hours prior to each interview. Based on these recalls, daily aggregated consumption of water, food energy, and other nutrition components such as total fat and total sugar consumption were computed.
In this application, we focus on the relationship between participants’ total cholesterol level (y), their age (Z), and their daily intakes of 42 nutrition components, such as sugars, total vitamins, fats, retinol, lycopene, zinc, and selenium, among many others. We restrict our analysis to \(n = 3343\) women, and assume participants’ ages are measured accurately while the daily intakes of nutrition components are subject to additive measurement errors. For the ith participant, let \(\textbf{W}_{i1}\) and \(\textbf{W}_{i2}\) denote the \(43 \times 1\) vector of surrogates at the first and second interview, respectively. For each vector, the first element is the age (\(Z_i\)) and the remaining elements are the recorded values for nutrition components at the corresponding interview time. We assume the classical measurement error model \(\textbf{W}_{ij} = \textbf{X}_i + \textbf{U}_{ij}\) for \(i=1,\ldots ,n\) and \(j=1,2\), where \(\textbf{X}_i\) denotes the vector of the long–term nutrition intakes, and \(\textbf{U}_i \sim N(\varvec{0}, {\varvec{\Sigma }}^*_u)\) denotes the measurement errors. Moreover, as \(\text {E}\left\{ (\textbf{W}_{i1}- \textbf{W}_{i2}) (\textbf{W}_{i1}- \textbf{W}_{i2})^{\text {T}} \right\} = 2{\varvec{\Sigma }}^*_u\), we estimate \(\widehat{{\varvec{\Sigma }}}^*_u = (2n)^{-1} \sum _{i=1}^{n} (\textbf{W}_{i1}- \textbf{W}_{i2}) (\textbf{W}_{i1}- \textbf{W}_{i2})^{\text {T}}\). Therefore, the covariance matrix of the measurement errors corresponding to \(\textbf{W}_i = (1/2)(\textbf{W}_{i1} + \textbf{W}_{i2})\) is estimated as \(\hat{\varvec{\Sigma }}_u = \widehat{\varvec{\Sigma }}_u^*/2\), which is a consistent estimator of \(\varvec{\Sigma }_u\) under general regularity conditions as \(n \rightarrow \infty \) (Carroll et al. 1993). Note since the age is assumed to be measured without error, the first row and column of \(\hat{\varvec{\Sigma }}_u\) are zero.
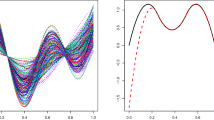
We estimate the central subspace \(\mathcal {S}_{y \vert \textbf{X}}\) using the three sparse estimators in the simulation studies, setting \(d=1\) and \(H=20\) slices. Letting \(\hat{\varvec{\beta }}^{(\text {IL-Tan})}\) and \(\hat{\varvec{\beta }}^{(\text {IL-Lin})}\) be the estimated bases from the IL-Tan and IL-Lin estimators, respectively, the sufficient predictors are estimated to be \(\hat{T}_i^{(\text {IL-Tan})} = \hat{\textbf{X}}_i\hat{\varvec{\beta }}^{(\text {IL-Tan})}\) and \(\hat{T}_i^{(\text {IL-Lin})}=\hat{\textbf{X}}_i\hat{\varvec{\beta }}^{(\text {IL-Lin})}\), where \(\hat{\textbf{X}}_i = (\hat{\varvec{\Sigma }}_w-\hat{\varvec{\Sigma }}_u)\hat{\varvec{\Sigma }}_w^{-1} \textbf{W}_i\). For the scLAD estimator \(\hat{\varvec{\beta }}^{(\text {scLAD})}\), we denote the corresponding sufficient predictor as \( \hat{T}_i^{(\text {scLAD})} = \hat{\textbf{V}}_i\hat{\varvec{\beta }}^{(\text {scLAD})}\), where \(\hat{\textbf{V}}_i = \hat{\varvec{\Delta }}(\hat{\varvec{\Delta }} + \hat{\varvec{\Sigma }}_u)^{-1}\textbf{W}_i\). Figure 1 presents the plots of y against the three sufficient predictors.
It can be seen that the sufficient predictors formed from scLAD and IL-Lin estimators are relatively similar to each other, and are more informative about the total cholesterol level than the IL-Tan estimator. Indeed, the sample correlation between the outcome and \(\hat{\textbf{T}}^{(\text {IL-Tan})}\) is 0.04, but the sample correlations between the outcome and \(\hat{\textbf{T}}^{(\text {IL-Lin})}\) and \(\hat{\textbf{T}}^{(\text {IL-scLAD})}\) are 0.45 and 0.43, respectively. In terms of variable selection, out of 43 predictors, the IL-Lin estimator selects 27 variables, the IL-Tan estimator selects 15 variables, while the proposed scLAD estimator selects 17 variables. The simulation results suggest that IL-Lin is potentially overfitting. Among the variables selected by scLAD, the three predictors that have the largest magnitudes are copper, vitamin B12, and zinc.
7 Discussion
In this article, we propose two likelihood-based estimators for the central subspace when the predictors are contaminated by additive measurement errors. These estimators are constructed from a Gaussian inverse regression model \(\textbf{X}\mid y\) into which measurement error is incorporated in a straightforward manner. The two estimators are based on maximizing the likelihood-based objective functions of two different adjusted covariates, the first one using the conditional covariance \(\text {E}\left\{ \text {Var}(\textbf{X}\mid y) \right\} \) and the second one using the marginal covariance of \(\textbf{X}\). We establish the asymptotic equivalence of these two estimators. When the number of covariates is large, we propose a sparse corrected LAD estimator that facilitates variable selection by introducing a penalty term that regularizes the projection matrix directly. The corresponding objective function is defined on an appropriate Grassmann manifold, and has closed-form gradients that facilitate efficient computation. Simulation studies generating data from forward index models demonstrate that the likelihood-based estimators have superior performance to the inverse moment-based estimators in terms of both estimation and variable selection consistency. Note that in our simulation studies, we did not consider generating data directly from the inverse regression model. We anticipate that the overall conclusions would be similar to, if not better than in terms of favoring our proposed estimators, those presented in this article with forward index models i.e., where we generated the covariate \(\textbf{X}\) from the marginal distribution and then generated the response y from the conditional distribution \(y \mid \textbf{B}^{\text {T}}op \textbf{X}\).
Future research can address the theoretical properties of the sparse likelihood-based estimators, particularly their variable selection consistency (see for instance Lin et al. 2019; Nghiem et al. 2022, for related works), and develop likelihood-based estimators of the central subspace that are robust to non-Gaussian measurement errors. Furthermore, we can consider extending the proposed method to the more general linear measurement error model, \(\textbf{W}= \textbf{A}\textbf{X}+ \textbf{U}\), with \(\textbf{A}\ne \textbf{I}_p\). We conjecture that our proposed estimators, along with their corresponding theoretical results, will still hold for this more general model (assuming \(\textbf{A}\) is either known or can be estimated consistently). Furthermore, the issue of estimating the number of dimensions d from the surrogate data remains open.
8 Supplementary materials
The online Supplementary Materials contain additional simulation results that are referred in Sect. 5. The MATLAB code implementing the corrected estimators is available at https://github.com/lnghiemum/LAD-ME.
References
Absil, P.-A., Baker, C.G., Gallivan, K.A.: Trust-region methods on Riemannian manifolds. Found. Comput. Math. 7, 303–330 (2007)
Boumal, N., Mishra, B., Absil, P.-A., Sepulchre, R.: Manopt, a Matlab toolbox for optimization on manifolds. J. Mach. Learn. Res. 15(42), 1455–1459 (2014)
Carroll, R.J., Li, K.-C.: Measurement error regression with unknown link: dimension reduction and data visualization. J. Am. Stat. Assoc. 87, 1040–1050 (1992)
Carroll, R.J., Eltinge, J.L., Ruppert, D.: Robust linear regression in replicated measurement error models. Stat. Probab. Lett. 16, 169–175 (1993)
Carroll, R.J., Ruppert, D., Stefanski, L.A., Crainiceanu, C.M.: Measurement Error in Nonlinear Models: A Modern Perspective. Chapman and Hall/CRC, Boca Raton, FL (2006)
Centers for Disease Control and Prevention: National Health and Nutrition Examination Survey Data. U.S. Department of Health and Human Services, Centers for Disease Control and Prevention (2022)
Chen, L.-P.: De-noising boosting methods for variable selection and estimation subject to error-prone variables. Stat. Comput. 33, 38 (2023)
Chen, L.-P., Yi, G.Y.: Sufficient dimension reduction for survival data analysis with error-prone variables. Electron. J. Stat. 16(1), 2082–2123 (2022). https://doi.org/10.1214/22-EJS1977
Cook, R.: SAVE: a method for dimension reduction and graphics in regression. Commun. Stat.-Theory Methods 29, 2109–2121 (2000)
Cook, R.D.: Regression Graphics: Ideas for Studying Regressions Through Graphics, vol. 482. Wiley, New York, NY (2009)
Cook, R.D., Forzani, L.: Principal fitted components for dimension reduction in regression. Stat. Sci. 23, 485–501 (2008)
Cook, R.D., Forzani, L.: Likelihood-based sufficient dimension reduction. J. Am. Stat. Assoc. 104, 197–208 (2009)
Gallivan, K.A., Srivastava, A., Liu, X., Van Dooren, P.: Efficient algorithms for inferences on Grassmann manifolds. In: IEEE Workshop on Statistical Signal Processing, 2003, pp. 315–318 (2003). IEEE
Glaws, A., Constantine, P.G., Cook, R.D.: Inverse regression for ridge recovery: a data-driven approach for parameter reduction in computer experiments. Stat. Comput. 30, 237–253 (2020)
Grace, Y.Y., Delaigle, A., Gustafson, P.: Handbook of Measurement Error Models. CRC Press, Boca Raton, FL (2021)
Hui, F.K.C., Nghiem, L.H.: Sufficient dimension reduction for clustered data via finite mixture modelling. Aust. N. Z. J. Stat. 64, 133–157 (2022)
Li, K.-C.: Sliced inverse regression for dimension reduction. J. Am. Stat. Assoc. 86, 316–327 (1991)
Li, B.: Sufficient Dimension Reduction: Methods and Applications with R. Chapman and Hall/CRC, Boca Raton, FL (2018)
Li, B., Wang, S.: On directional regression for dimension reduction. J. Am. Stat. Assoc. 102, 997–1008 (2007)
Li, B., Yin, X.: On surrogate dimension reduction for measurement error regression: an invariance law. Ann. Stat. 35, 2143–2172 (2007)
Lin, Q., Zhao, Z., Liu, J.S.: Sparse sliced inverse regression via lasso. J. Am. Stat. Assoc. 114, 1–33 (2019). https://doi.org/10.1080/01621459.2018.1520115
Lin, Q., Zhao, Z., Liu, J.S.: Sparse sliced inverse regression via lasso. J. Am. Stat. Assoc. 114, 1726–1739 (2019)
Lin, Q., Li, X., Huang, D., Liu, J.S.: On the optimality of sliced inverse regression in high dimensions. Ann. Stat. 49(1), 1–20 (2021). https://doi.org/10.1214/19-AOS1813
Nghiem, L., Hui, F.K.C., Muller, S., Welsh, A.H.: Sparse sliced inverse regression via Cholesky matrix penalization. Stat. Sin. 32, 2431–2453 (2022). https://doi.org/10.5705/ss.202020.0406
Nghiem, L.H., Hui, F.K.C., Müller, S., Welsh, A.H.: Sparse sliced inverse regression via Cholesky matrix penalization. Stat. Sin. 32, 2431–2453 (2022)
Nghiem, L.H., Hui, F.K.C., Müller, S., Welsh, A.H.: Screening methods for linear errors-in-variables models in high dimensions. Biometrics 79, 926–939 (2023)
Qian, W., Ding, S., Cook, R.D.: Sparse minimum discrepancy approach to sufficient dimension reduction with simultaneous variable selection in ultrahigh dimension. J. Am. Stat. Assoc. 114, 1277–1290 (2019)
Rao, C.R., Rao, C.R., Statistiker, M., Rao, C.R., Rao, C.R.: Linear Statistical Inference and Its Applications, vol. 2. Wiley, New York, NY (1973)
Reich, B.J., Bondell, H.D., Li, L.: Sufficient dimension reduction via Bayesian mixture modeling. Biometrics 67, 886–895 (2011)
Tan, K.M., Wang, Z., Zhang, T., Liu, H., Cook, R.D.: A convex formulation for high-dimensional sparse sliced inverse regression. Biometrika 105, 769–782 (2018)
Wang, Q., Gao, J., Li, H.: Grassmannian manifold optimization assisted sparse spectral clustering. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 5258–5266 (2017)
Xia, Y., Tong, H., Li, W.K., Zhu, L.-X.: An adaptive estimation of dimension reduction space. J. R. Stat. Soc. Ser. B (Stat. Methodol.) 64, 363–410 (2002)
Zhang, J., Zhu, L.-P., Zhu, L.-X.: On a dimension reduction regression with covariate adjustment. J. Multivar. Anal. 104, 39–55 (2012)
Zhang, J., Zhu, L., Zhu, L.: Surrogate dimension reduction in measurement error regressions. Stat. Sin. 24, 1341–1363 (2014)
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. FKCH, SM, and AHW were supported by Australian Research Council Discovery Grants DP230101908.
Author information
Authors and Affiliations
Contributions
Nghiem initiated the ideas and implemented the methods. All authors contributed to the ideas, wrote the main manuscript text, and reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Appendices
Appendices
1.1 Appendix A: Proof of Proposition 1
Throughout the proof, we use the following equalities from Cook and Forzani (2009), p. 205, which are from Rao et al. (1973), p. 77. Let \(\textbf{B}\in \mathbb {R}^{p\times p}\) be a symmetric positive definite matrix, and \((\varvec{\Psi }, \varvec{\Psi }_0) \in \mathbb {R}^{p \times p}\) be an orthogonal matrix. Then we have
Recall that \(\textbf{V}= \textbf{L}\textbf{W}\) with \(\textbf{L}= \varvec{\Delta }(\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1}\), and the inverse Gaussian model \(\textbf{X}\mid y \sim N(\varvec{\mu }_y, \varvec{\Delta }_y)\) with \(\varvec{\mu }_y - \varvec{\mu }= \varvec{\Delta }\varvec{\Psi }\varvec{v}_y\) and \(\varvec{\Psi }_0 \varvec{\Delta }_y^{-1} = \varvec{\Psi }_0 \varvec{\Delta }^{-1}\). The joint distribution of \(\varvec{\Psi }^{\text {T}} \textbf{V}\) and \(\varvec{\Psi }_0^{\text {T}} \textbf{V}\) given y is
where
with \(\textbf{G}_y = \textbf{L}(\varvec{\Delta }_y + \varvec{\Sigma }_u) \textbf{L}^{\text {T}} = \varvec{\Delta }(\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1} (\varvec{\Delta }_y + \varvec{\Sigma }_u) (\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1} \varvec{\Delta }\). Hence part (i) of Proposition 1 follows. For part (ii), the conditional distribution \(\varvec{\Psi }_0^{\text {T}} \textbf{V}\mid (\varvec{\Psi }^{\text {T}} \textbf{V}, y)\) is normal. The conditional mean is
This conditional mean does not depend on y; indeed, the coefficient associated with \(\varvec{v}_y\) equals
To see the last equality, applying (7) with \(\textbf{B}= \textbf{G}_y\), we have \( (\varvec{\Psi }^{\text {T}} \textbf{G}_y \varvec{\Psi }_0)(\varvec{\Psi }^{\text {T}} \textbf{G}_y \varvec{\Psi })^{-1} = - (\varvec{\Psi }_0^{\text {T}} \textbf{G}_y^{-1} \varvec{\Psi }_0)^{-1} (\varvec{\Psi }_0^{\text {T}} \textbf{G}_y^{-1}\varvec{\Psi }), \) and furthermore,
where steps (a) and (b) follow from \(\varvec{\Psi }_0^{\text {T}} \varvec{\Delta }_y^{-1} = \varvec{\Psi }_0^{\text {T}} \varvec{\Delta }^{-1}\). Therefore,
where the last equality follows from applying (7) with \(\textbf{B}= \textbf{L}\varvec{\Delta }\). Substituting it into the left hand side of (12) gives the claim. Hence the conditional expectation in (11) is reduced to \( \text {E}\left( \varvec{\Psi }_0^{\text {T}} \textbf{V}\vert \varvec{\Psi }^{\text {T}} \textbf{V}, y \right) = \varvec{\Psi }_0^{\text {T}} \textbf{L}\varvec{\mu }+ (\varvec{\Psi }_0^{\text {T}} \textbf{L}\varvec{\Delta }\varvec{\Psi })(\varvec{\Psi }^{\text {T}} \textbf{L}\varvec{\Delta }\varvec{\Psi })^{-1}(\varvec{\Psi }^{\text {T}} \textbf{L}\textbf{W}- \varvec{\Psi }^{\text {T}} \textbf{L}\varvec{\mu }) = (\varvec{\Psi }_0^{\text {T}} - \textbf{H}\varvec{\Psi }^{\text {T}}) \textbf{L}\varvec{\mu }+ \textbf{H}\varvec{\Psi }^{\text {T}} \textbf{L}\textbf{W}\). Next, the conditional covariance matrix of \(\varvec{\Psi }_0^{\text {T}} \textbf{V}\) given \(\varvec{\Psi }^{\text {T}} \textbf{V}\) and y equals
which does not depend on y either, where the second equality follows from (8) with \(\textbf{B}= \textbf{G}_y\). The proof is hence complete.
1.2 Appendix B: Proof of Theorem 1
We estimate the central subspace of \(\mathcal {S}_{y \vert \textbf{X}}\) by maximizing the joint log likelihood of \(\varvec{\Psi }^{\text {T}} \textbf{V}_i^{(m)}\) and \(\varvec{\Psi }_0^{\text {T}} \textbf{V}_i^{(m)}\) for \(i=1,\ldots , n\) and \(m=1,\ldots , M\). Let \(n_m\) denote the number of observations in the jth slice, and \(f_m = n_m/n\). Let \(\overline{\textbf{W}}_m\) and \(\tilde{\varvec{\Delta }}_{wm}\) denote the sample mean and sample covariance of the surrogate data within the mth slice, respectively. Let \(\textbf{G}_m = \textbf{L}(\varvec{\Delta }_y^{(m)} + \varvec{\Sigma }_u) \textbf{L}^{\text {T}} = \varvec{\Delta }(\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1} (\varvec{\Delta }_y^{(m)} + \varvec{\Sigma }_u) (\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1} \varvec{\Delta }\). For any square matrix \(\textbf{A}\), we use \({\text {tr}}(\textbf{A})\) to denote its trace. Then, the joint log-likelihood of the data is given by
with \( \textbf{K}:=\varvec{\Psi }_0 - \varvec{\Psi }\textbf{H}^{\text {T}}\). We maximize the log likelihood with the two constraints that \(\sum _{m=1}^{M} f_m \varvec{v}_m = 0\), and \(\varvec{\Psi }^{\text {T}} \left( \sum _{m=1}^{M} f_m \textbf{G}_m \right) \varvec{\Psi }= \varvec{\Psi }^{\text {T}} \textbf{L}\varvec{\Delta }\varvec{\Psi }\). First, we maximize with respect to \(\varvec{v}_m\); only the third term in the log-likelihood above contains \(\varvec{v}_m\) so combining this term with the first constraint, we need to find \(\varvec{v}_m\) that minimizes
where \(\textbf{Z}_m :=\varvec{\Psi }^{\text {T}} \textbf{L}(\overline{\textbf{W}}_m - \varvec{\mu })\), \(\textbf{B}:=\varvec{\Psi }^{\text {T}} \textbf{L}\varvec{\Delta }\varvec{\Psi }\), and \(\textbf{B}_m :=\varvec{\Psi }^{\text {T}} \textbf{G}_m \varvec{\Psi }\). Differentiating (14) with respect to \(\varvec{v}_m\) and setting the derivatives equal zero, we obtain \(2 f_m\textbf{B}\textbf{B}_m^{-1}(\textbf{Z}_m - \textbf{B}\varvec{v}_m) + \lambda f_m = 0\), or equivalently, \(2f_m \textbf{Z}_m - \textbf{B}f_m\varvec{v}_m + f_m\textbf{B}_m\textbf{B}^{-1}\lambda = 0 \). Summing over all m and using the constraints, we have \(\lambda = \sum _{m=1}^{M} f_m \textbf{Z}_m :=\bar{\textbf{Z}}\), and hence \(\textbf{Z}_m - \textbf{B}\varvec{v}_m = \textbf{B}_m \textbf{B}^{-1} \lambda \). Therefore, at the optimized value of \(\varvec{v}_m\), (14) reduces to
with \(\overline{\textbf{W}} = \sum _{m=1}^{M} f_m \overline{\textbf{W}}_m\).
Next, we maximize the log-likelihood with respect to \(\varvec{\mu }\) at the optimized value of \(\varvec{v}_m\). Taking the derivative with respect to \(\varvec{\mu }\) and setting it equal to zero, we obtain
Noting that
so we obtain
Furthermore, \(\varvec{\Psi }\textbf{B}^{-1} \varvec{\Psi }^{\text {T}} = \varvec{\Psi }(\varvec{\Psi }^{\text {T}} \textbf{L}\varvec{\Delta }\varvec{\Psi })^{-1} \varvec{\Psi }^{\text {T}}\), so using (9) with \(\textbf{B}= \textbf{L}\varvec{\Delta }\), we obtain
Therefore, Eq. (15) reduces to \( \textbf{L}^{\text {T}}(\textbf{L}\varvec{\Delta })^{-1}\textbf{L}(\overline{\textbf{W}}-\varvec{\mu }) = \textbf{L}^{\text {T}} \varvec{\Delta }^{-1}(\overline{\textbf{W}}-\varvec{\mu }) = 0. \) The matrix \(\textbf{L}^{\text {T}} \varvec{\Delta }^{-1} = (\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1} \varvec{\Delta }\varvec{\Delta }^{-1} = (\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1}\) is invertible, so the above equation gives the optimized value \(\hat{\varvec{\mu }} = \overline{\textbf{W}}\). At these optimized values \(\hat{\varvec{\mu }}\) and \(\hat{v}_m\), the log-likelihood is reduced to
with \(\tilde{\varvec{\Sigma }}_m = \tilde{\varvec{\Delta }}_{wm} + (\overline{\textbf{W}}_m - \overline{\textbf{W}}) (\overline{\textbf{W}}_m - \overline{\textbf{W}})^{\text {T}}\). Next, let \(\textbf{S}_m = \varvec{\Psi }^{\text {T}} \textbf{G}_m \varvec{\Psi }\), such that we can write
We maximize \(\ell _d\) with respect to \(\textbf{S}_m\). Taking the derivative of \(\ell _d\) with respect to \(\textbf{S}_m\) and setting it equal to zero, we obtain \(\hat{\textbf{S}}_m - \varvec{\Psi }^{\text {T}} \textbf{L}\tilde{\varvec{\Delta }}_{wm} \textbf{L}^{\text {T}} \varvec{\Psi }= 0\), from which we obtain \(\hat{\textbf{S}}_m = \varvec{\Psi }^{\text {T}} \textbf{L}\tilde{\varvec{\Delta }}_{wm} \textbf{L}^{\text {T}} \varvec{\Psi }\) (see the last line of page 206 in Cook and Forzani 2009, for a similar step). At this optimized value \(\hat{\textbf{S}}_m\), we obtain
Next, we find the maximum likelihood of \(\textbf{H}\). Since \(\textbf{K}= \varvec{\Psi }_0 - \varvec{\Psi }\textbf{H}^{\text {T}}\), the derivative with respect to \(\textbf{H}\) equals \(-\sum _{m=1}^{M} n_m \textbf{D}^{-1} \varvec{\Psi }_0^{\text {T}} \textbf{L}\tilde{\varvec{\Sigma }}_m \textbf{L}^{\text {T}}\varvec{\Psi }+ \sum _{m=1}^{M} n_m \textbf{D}^{-1}\textbf{H}\varvec{\Psi }^{\text {T}} \textbf{L}\tilde{\varvec{\Sigma }}_m \textbf{L}^{\text {T}} \varvec{\Psi }, \) which gives the maximum at \( \hat{\textbf{H}} = (\varvec{\Psi }_0^{\text {T}} \textbf{L}\tilde{\varvec{\Sigma }} \textbf{L}^{\text {T}} \varvec{\Psi }) (\varvec{\Psi }^{\text {T}} \textbf{L}\tilde{\varvec{\Sigma }} \textbf{L}^{\text {T}} \varvec{\Psi })^{-1} \) where \(\tilde{\varvec{\Sigma }} :=\sum _{m=1}^{M} f_m \tilde{\varvec{\Sigma }}_m\). Hence, \(\hat{\textbf{K}}^{\text {T}} = \varvec{\Psi }_0^{\text {T}} - \hat{\textbf{H}}\varvec{\Psi }^{\text {T}} = \varvec{\Psi }_0^{\text {T}} (\textbf{I}_p - \textbf{L}\tilde{\varvec{\Sigma }} \textbf{L}^{\text {T}} \varvec{\Psi }(\varvec{\Psi }^{\text {T}} \textbf{L}\tilde{\varvec{\Sigma }} \textbf{L}^{\text {T}} \varvec{\Psi })^{-1}) = \{\varvec{\Psi }_0^{\text {T}} (\textbf{L}\tilde{\varvec{\Sigma }} \textbf{L}^{\text {T}})^{-1}\varvec{\Psi }_0\}^{-1} \varvec{\Psi }_0^{\text {T}} (\textbf{L}\tilde{\varvec{\Sigma }} \textbf{L}^{\text {T}})^{-1} \), and the maximum over \(\textbf{D}\) is
Finally, substituting \(\hat{\textbf{D}}\) into (17) and removing terms that do not involve \(\varvec{\Psi }\) and \(\textbf{L}\), we obtain the joint profile likelihood of \(\varvec{\Psi }\) and \(\varvec{\Delta }\) to be
1.3 Appendix C: Proof of Theorem 2
We begin by establishing the equivalence between \(\mathcal {S}^*_{\text {cLAD}}\) and \(\mathcal {S}^*_{\text {SAVE}}\). First, we establish the equivalence when \(\varvec{\Delta }= \text {E} \left\{ \text {Var}(\textbf{X}\mid y) \right\} \) (and hence \(\textbf{L}\)) is known, and then we will establish the consistency of the method of moment estimator \(\hat{\varvec{\Delta }}\). To simplify the notation, we will use \(\varvec{\Delta }_m\) to denote \(\varvec{\Delta }_y^{(m)}\), i.e., the covariance matrix of the inverse Gaussian distribution within the mth slice.
Let \(\mathcal {V}_1(\varvec{\Psi }) = n \log \vert \varvec{\Psi }^{\text {T}} (\textbf{L}\tilde{\varvec{\Sigma }}_w \textbf{L}^{\text {T}}) \varvec{\Psi }\vert - \sum _{m=1}^{M} n_m \log \vert \varvec{\Psi }^{\text {T}} \textbf{L}\tilde{\varvec{\Delta }}_{wm} \textbf{L}^{\text {T}} \varvec{\Psi }\vert \), where \(\varvec{\Psi }\in \mathbb {R}^{p \times d}\) is a semi-orthogonal matrix. When \(n \rightarrow \infty \), the function \(n^{-1} \mathcal {V}_1(\varvec{\Psi })\) converges uniformly to \(\mathcal {V}_0(\varvec{\Psi }) = \log \vert \varvec{\Psi }^{\text {T}} (\textbf{L}\varvec{\Sigma }_w \textbf{L}^{\text {T}}) \varvec{\Psi }\vert - \sum _{m=1}^{M} f_m \log \vert \varvec{\Psi }^{\text {T}} (\textbf{L}\varvec{\Delta }_{wm} \textbf{L}^{\text {T}}) \varvec{\Psi }\vert \), where \(\varvec{\Delta }_{wm} = \varvec{\Delta }_m + \varvec{\Sigma }_u\) and \(\varvec{\Sigma }_w = \varvec{\Sigma }_x + \varvec{\Delta }_u\). Using the result from Cook (2009) that \( \vert \varvec{\Psi }_0^{\text {T}} \textbf{B}\varvec{\Psi }_0 \vert = \vert \textbf{B}\vert \vert \varvec{\Psi }^{\text {T}} \textbf{B}^{-1} \varvec{\Psi }\vert \) for any symmetric positive semidefinite matrix \(\textbf{B}\), we obtain
where c is a quantity that does not depend on \(\varvec{\Psi }\) or \(\varvec{\Psi }_0\). To show the consistency of the estimated central subspace, we show that the true SAVE estimator (i.e defined even when no measurement error is present) is the global maximum for \(\mathcal {V}_0(\varvec{\Psi })\).
Indeed, as a function of Q, the function \(\log \vert \varvec{\Psi }_0^{\text {T}} Q^{-1} \varvec{\Psi }_0 \vert \) is convex. As a result, we obtain
where the second inequality follows from \(\sum _{m=1}^{M} f_m = 1\) and \(\sum _{m=1}^{M} f_m \varvec{\Delta }_m = \varvec{\Delta }\). Next we note that \(\varvec{\Sigma }_x = \varvec{\Delta }+ \text {Var}\left\{ \text {E}(\textbf{X}|y)\right\} \), so \(\varvec{\Sigma }_x + \varvec{\Sigma }_u - (\varvec{\Delta }_x + \varvec{\Sigma }_u) = \varvec{\Sigma }_x - \varvec{\Delta }\) is still a positive definite matrix. As a result, the matrix difference \( (\varvec{\Sigma }_x + \varvec{\Sigma }_u)^{-1} - (\varvec{\Delta }+ \varvec{\Sigma }_u)^{-1}\) is negative definite, and so is \(\varvec{\Psi }_0^{\text {T}} \left\{ \textbf{L}(\varvec{\Sigma }_x +\varvec{\Sigma }_u)\textbf{L}^{\text {T}} \right\} ^{-1} \varvec{\Psi }_0- \varvec{\Psi }_0^{\text {T}} \left\{ \textbf{L}(\varvec{\Delta }+\varvec{\Sigma }_u)\textbf{L}^{\text {T}} \right\} ^{-1}\varvec{\Psi }_0\). Therefore, we have
As a result, we have \(\mathcal {V}_0(\varvec{\Psi }) \le c\) and the equality holds only when
We next prove that if \({\varvec{\Phi }}\) is a semi-orthogonal basis matrix for the population SAVE subspace when \(\textbf{X}\) is observed and \(({\varvec{\Phi }}, {\varvec{\Phi }}_0) \in \mathbb {R}^{p \times p}\) is an orthogonal matrix (i.e \({\varvec{\Phi }}^{\text {T}} {\varvec{\Phi }}_0 = 0\)), then Eq. (19) is satisfied with \(\varvec{\Psi }_0\) replaced by \({\varvec{\Phi }}_0\). Cook and Forzani (2009) prove that \({\varvec{\Phi }}\) satisfies \(\varvec{\Delta }_m^{-1} = \varvec{\Sigma }^{-1} + {\varvec{\Phi }} \left\{ ({\varvec{\Phi }}^{\text {T}} \varvec{\Delta }_m {\varvec{\Phi }})^{-1} - ({\varvec{\Phi }}^{\text {T}} \varvec{\Sigma }{\varvec{\Phi }})^{-1}\right\} {\varvec{\Phi }}^{\text {T}}\); as a result, we have \({\varvec{\Phi }}_0^{\text {T}} \varvec{\Delta }_{m}^{-1} = {\varvec{\Phi }}_0^{\text {T}} \varvec{\Sigma }^{-1}\). Furthermore, asymptotically, Cook and Forzani (2009) prove that when \(\textbf{X}\) is observed, then LAD and SAVE estimate the same subspace, and hence \({\varvec{\Phi }}_0^{\text {T}} \varvec{\Delta }_m^{-1} = {\varvec{\Phi }}_0^{\text {T}} \varvec{\Delta }^{-1}\) as well. As a result, we have
and
which verifies (19) for the population SAVE subspace.
Finally, since we estimate \(\textbf{L}\) by \(\hat{\textbf{L}}\) from the naive LAD estimator (see the paragraph above Eq. eqrefeq:MLE2 for the definition of the naive LAD estimator), we will prove that \(\hat{\textbf{L}}\) is consistent for \(\textbf{L}\). It suffices to prove that \(\hat{\varvec{\Delta }}_{\text {n}}\) is a consistent estimator of \(\text {E}\left\{ \text {Var}(\textbf{W}\mid y\right\} \). By the relationship between LAD and SAVE, \(\hat{\varvec{\Psi }}_{\text {n}}\) converges to a naive SAVE population subspace, formally defined as \(\mathcal {S}_{\text {n}} = \Sigma _w^{-1} \text {span}\{{\varvec{\Sigma }}_w - ({\varvec{\Delta }}_1 + {\varvec{\Sigma }}_u), , \ldots , {\varvec{\Sigma }}_w - ({\varvec{\Delta }}_m + {\varvec{\Sigma }}_u) = \Sigma _w^{-1} \text {span}\{{\varvec{\Sigma }}_x - {\varvec{\Delta }}_1, , \ldots , {\varvec{\Sigma }}_x - {\varvec{\Delta }}_m \}\) with a semi-orthogonal basis \(\Phi _{\text {n}}\) satisfying
and
Therefore, as \(n \rightarrow \infty \), we have
converges to
so \(\hat{\varvec{\Delta }}_n\) converges to \(\varvec{\Sigma }_u + \varvec{\Delta }= \text {E}\left\{ \text {Var}(\textbf{W}\mid y) \right\} \). The proof is now complete.
To prove the equivalence between \(\mathcal {S}^*_{\text {IL--LAD}}\) and \(\mathcal {S}_{\text {SAVE}}\), by a similar argument, it suffices to establish
where \({\varvec{\Phi }}_0\) is the orthogonal complement of the semi-orthogonal basis \({\varvec{\Phi }}\) corresponding to the population SAVE subspace, and with \({\varvec{\Sigma }}^* = {\varvec{\Sigma }}_x{\varvec{\Sigma }}_w^{-1}{\varvec{\Sigma }}_x\), and \({\varvec{\Delta }}_m^* = {\varvec{\Sigma }}_x {\varvec{\Sigma }}_{w}^{-1} ( {\varvec{\Delta }}_m+ {\varvec{\Sigma }}_u){\varvec{\Sigma }}_{w}^{-1}{\varvec{\Sigma }}_x\). Since \({\varvec{\Phi }}_0^{\text {T}} {\varvec{\Sigma }}_x^{-1} = {\varvec{\Phi }}_0^{\text {T}} {\varvec{\Delta }}_m^{-1}\), we have
so (20) follows from \(\sum _{m=1}^{M} f_m = 1\).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nghiem, L.H., Hui, F.K.C., Muller, S. et al. Likelihood-based surrogate dimension reduction. Stat Comput 34, 51 (2024). https://doi.org/10.1007/s11222-023-10357-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11222-023-10357-6