Abstract
The spatio-temporal epidemic type aftershock sequence (ETAS) model is widely used to describe the self-exciting nature of earthquake occurrences. While traditional inference methods provide only point estimates of the model parameters, we aim at a fully Bayesian treatment of model inference, allowing naturally to incorporate prior knowledge and uncertainty quantification of the resulting estimates. Therefore, we introduce a highly flexible, non-parametric representation for the spatially varying ETAS background intensity through a Gaussian process (GP) prior. Combined with classical triggering functions this results in a new model formulation, namely the GP-ETAS model. We enable tractable and efficient Gibbs sampling by deriving an augmented form of the GP-ETAS inference problem. This novel sampling approach allows us to assess the posterior model variables conditioned on observed earthquake catalogues, i.e., the spatial background intensity and the parameters of the triggering function. Empirical results on two synthetic data sets indicate that GP-ETAS outperforms standard models and thus demonstrate the predictive power for observed earthquake catalogues including uncertainty quantification for the estimated parameters. Finally, a case study for the l’Aquila region, Italy, with the devastating event on 6 April 2009, is presented.
Similar content being viewed by others
Explore related subjects
Find the latest articles, discoveries, and news in related topics.Avoid common mistakes on your manuscript.
1 Introduction
Point process models are often used in statistical seismology for describing the occurrence of earthquakes (point data) in a spatio-temporal setting. The most widely used one is the epidemic type aftershock sequence (ETAS) model, first introduced as a temporal point process model (Ogata 1988), and later enhanced to the currently predominantly employed spatio-temporal version (Ogata 1998). Main applications are seismic forecasting or the characterisation of earthquake clustering in a particular geographical region and topics alike (e.g., Jordan et al. 2011). The ETAS model is an example of a self-exciting, spatio-temporal, marked point process, which is a particular Hawkes process model, extending the temporal Hawkes process (Hawkes 1971). Self-excitation means that one event can trigger a series of subsequent follow-up events (offspring), as in the case of earthquakes, main shocks and aftershocks. The ETAS model as a variant of a Hawkes process model assigns the earthquake magnitude as a mark to each event, and it usually employs a specific mark depending excitation kernel (Ogata 1988, 1998). Besides its primary application in seismology, the Hawkes process is utilised in several other domains, e.g. finance (Bacry et al. 2015; Filimonov and Sornette 2015), crime (Porter and White 2010; Mohler et al. 2011), neuronal activities (Gerhard et al. 2017), social networks (Zhou et al. 2013; Zhao et al. 2015), genomes (Reynaud-Bouret and Schbath 2010), transportation (Hu and Jin 2017).
The ETAS model is characterized by its conditional intensity function, that is, the rate of arriving events conditioned on the history of previous events. This time-dependent conditional intensity function itself consists of two parts, (i) a background intensity \(\mu \) of a Poisson process, which models the arrival of spontaneous (exogenous) events, and (ii) a time-dependent triggering function \(\varphi \) which encodes the form of self-excitation by adding a positive impulse response for each event, that is, an instantaneous jump which decays gradually at time progresses. An alternative approach interprets the stationary Hawkes process (e.g. the ETAS model) as a Poisson cluster process or branching process (Hawkes and Oakes 1974), which leads to the concept of a non-observable, underlying branching structure (a latent variable). One assumes that each event is either (i) a spontaneous, exogenous background event, or (ii) an offspring event, i.e., it was triggered by an existing event. Background events are generated independently by a Poisson process with rate \(\mu \) and form cluster centers. Each event may produce offspring, i.e., may become a parent of triggered events in the future, where each triggered event may produce further offspring. The intensity of these offspring processes is controlled by the triggering function \(\varphi \). Thus, each event has either a direct parent from which it was generated (offspring events) or is background (exogenous events with no direct parent); this yields an ordered branching structure useful for designing simulation and inference algorithms, e.g. (Zhuang et al. 2002; Veen and Schoenberg 2008).
The fitting of an ETAS model to data entails learning the conditional intensity function. Most currently used ETAS models employ a parametric form for the background \(\mu \) and the triggering function \(\varphi \). The parameters are then calibrated via maximum-likelihood estimation (MLE), maximising the classical likelihood function for point processes. Unfortunately, MLE has no simple analytical form. Alternatively, different numerical optimisation methods are employed involving, e.g., an Expectation-Maximisation (EM; Dempster et al. 1977) algorithm using the latent branching structure (Ogata 1998; Veen and Schoenberg 2008; Lippiello et al. 2014; Lombardi 2015).
Non-parametric methods have also been suggested previously to fit the conditional intensity function (or parts of it). For example, Zhuang et al. (2002) and Adelfio and Chiodi (2014) fit simultaneously a non-parametric background intensity via kernel density estimation and a classical parametric triggering kernel; Marsan and Lengliné (2008) consider a constant background intensity combined with a non-parametric histogram estimator of the triggering kernel; Mohler et al. (2011) suggest non-parametric kernel density estimators for both the components, background \(\mu \) and offspring \(\varphi \); and Fox et al. (2016) propose a non-parametric kernel density estimator for the background and non-parametric histogram estimation for the triggering kernel. Furthermore, Bacry and Muzy (2016) suggest a non-Bayesian, non-parametric way of estimating the triggering function of a Hawkes process based on Wiener Hopf integral equation; Kirchner (2017) presents a non-Bayesian non-parametric estimation procedure for a multivariate Hawkes process based on an integer-valued autoregressive model.
Uncertainty quantification of the ETAS model remains challenging. Most estimation techniques deliver a point estimate for its conditional intensity function and uncertainty quantification is usually achieved by relying on standard errors of estimated ETAS parameters, based on the Hessian (Ogata 1978; Rathbun 1996; Wang et al. 2010). This approach requires that the observational window is long enough (sufficiently large sample size), otherwise it may lead to an underestimation of parameter uncertainties. Moreover, standard errors based on Hessians cannot be obtained in the non-parametric case. Another approach to uncertainty quantification relies on various bootstrap techniques based on many forward simulations, e.g., Fox et al. (2016). Ad hoc variants for quantifying uncertainty have also been devised, e.g., by the solutions of multiple optimisation runs of the MLE, e.g. Lombardi (2015).
None of the aforementioned uncertainty quantification methods are fully satisfactory and we believe that a fully semi-parametric Bayesian framework is worthwhile pursuing, which allows one to incorporate prior knowledge. The posterior distribution effectively encodes the uncertainty of the quantities arising from data and a prior distribution. However, this poses a challenge for a spatio-temporal ETAS model, as there is no known conjugate structure, that is, the posterior can not be obtained in closed-form. One way to deal with this problem is to employ Monte Carlo sampling techniques, e.g. via Markov chain Monte Carlo (MCMC). However implementing MCMC remains challenging for non- or semi-parametric conditional intensity functions. Several studies have suggested Bayesian methods for the temporal, or multivariate Hawkes process, either based on parametric forms of the conditional intensity function (Rasmussen 2013; Ross 2018) or for non-parametric versions (Linderman and Adams 2015; Donnet et al. 2018; Zhang et al. 2019a, b; Zhou et al. 2019). But these studies rarely consider the spatio-temporal ETAS model and only with strong simplifications, e.g., a constant background intensity \(\mu \) (Rasmussen 2013). Recently, however, Kolev and Ross (2020) considered an inhomogeneous background intensity modelled via a Dirichlet process.
It is desirable to estimate the spatially dependent background \(\mu \) of an ETAS model fully non-parametrically as it is often difficult to specify an appropriate functional form a priori. The background intensity (also called long-term component) is of particular importance for seismic hazard assessment and seismic forecasting. It is often preferred to maintain a specific parametric triggering function \(\varphi \) (e.g., modified Omori law Omori 1894; Utsu 1961) as there is a long tradition for interpreting and comparing this particular parametric form in different settings, regions, etc. Thus, one faces two main issues for the development of a suitable Bayesian inference approach:
-
(i)
providing a Bayesian non-parametric way of modelling the background intensity \(\mu \), and
-
(ii)
creating a fully Bayesian inference algorithm for the resulting ETAS models including its parametric triggering component \(\varphi \).
We address these two issues in this paper by first formulating a Bayesian non-parametric approach to the estimation of the background intensity \(\mu \) via a Gaussian process (GP) prior. Secondly, we propose and implement a computationally tractable approach for the implied Bayesian inference problem by introducing auxiliary variables: a latent branching structure, a latent Poisson process, and latent Pólya–Gamma random variables. More specifically, we suggest to model the background intensity \(\mu \) non-parametrically by sigmoid transformed realisations of a GP prior, i.e., as a Sigmoid-Gauss-Cox-Process (SGCP; Adams et al. 2009), which is a doubly stochastic Poisson process. No specific functional form has to be chosen for the intensity function, and the prior fully specifies the chosen GP. Adams et al. (2009) proposed a Bayesian inference scheme via MCMC for SGCPs. However, the suggested scheme is computationally demanding and convergence is slow. Our paper relies instead on the work of Donner and Opper (2018) who recently enhanced Bayesian inference for SGCPs substantially by data augmentation with Pólya–Gamma random variables (Polson et al. 2013). The triggering function \(\varphi \) is modelled in a classical parametric way, which together with the SGCP model for \(\mu \) leads to a novel semi-parametric ETAS model formulation, which we denote as GP-ETAS. In order to implement such an approach, we need to address a number of computational challenges:
-
(i)
the background intensity \(\mu \) and the triggering function \(\varphi \) are not directly separable in the likelihood;
-
(ii)
intractable integrals for the posterior computation when \(\mu \) is modelled as SGCP, and
-
(iii)
handling a non-Gaussian point process likelihood while using a Gaussian process prior.
We show how these challenges can be resolved by data augmentation (introducing auxiliary variables), which strongly simplifies the Bayesian inference problem. It effectively allows us to construct an efficient MCMC sampling scheme for the posterior involving an overall Gibbs sampler (Geman and Geman 1984) consisting of three main steps, each conditioned on the other two:
-
(a)
conditionally sampling the latent branching structure which factorises the likelihood function into background and triggering component;
-
(b)
conditionally sampling the posterior of the background intensity \(\mu \) from explicit conditional densities easy to sample from; and
-
(c)
conditionally sampling the parameters of the triggering function \(\varphi \) by employing Metropolis–Hastings (MH; Hastings 1970) steps.
The remainder of this paper is structured as follows: First we describe the classical spatio-temporal ETAS model; secondly we introduce our GP-ETAS model including a simulation algorithm; thirdly the Bayesian inference approach is presented; fourthly empirical results based on synthetic and real data illustrate practical aspects of the framework. The paper concludes with a discussion and some final remarks.
2 Background
We start with a review of the classical spatio-temporal ETAS model, which we will use as a benchmark for comparison.
2.1 Classical ETAS model
The ETAS model (Ogata 1998), describes a stochastic process, which generates point pattern over some domain \(\mathcal {X}\times \mathcal {T}\times \mathcal {M}\), where \(\mathcal {T}\times \mathcal {X}\) is the time-space window and \(\mathcal {M}\) the mark space of the process. Realisations of this point process are denoted by \(\mathcal {D}=\{(t_i,\varvec{x}_i,m_i)\}_{i=1}^{N_{\mathcal {D}}}\), which in seismology can be interpreted as an earthquake catalog consisting of \(N_{\mathcal {D}}\) observed events. \(\mathcal {D}\) is usually ordered in time (time series), \(t_i \in \mathcal {T}\subseteq \mathbb {R}_{> 0}\) is the time of the ith event (time of the earthquake), \(\varvec{x}_i \in \mathcal {X}\subseteq \mathbb {R}^2\) is the corresponding location (longitude and latitude of the epicenter), and \(m_i \in \mathcal {M} \subseteq \mathbb {R}\) the corresponding mark (the magnitude of the earthquake).
2.1.1 Interpretations
There are two equivalent interpretations of the ETAS model (Hawkes process). We briefly discuss both.
Conditional intensity function. One way to define the ETAS model is by a conditional intensity function, which models the infinitesimal rate of expected arrivals around \((t,\varvec{x})\) given the history \(H_t=\{(t_i,\varvec{x}_i,m_i): t_i<t\}\) of the process until time t. The marked point process has intensity \(\tilde{\lambda }(t,\varvec{x},m|H_t) =\lambda (t,\varvec{x}|H_t)p_M(m)\), which factorizes under usual assumptions for the ETAS model (e.g., Zhuang et al. 2002; Daley and Vere-Jones 2003) in a ground process \(\lambda (t,\varvec{x}|H_t)\) given in (1) and a mark distribution \(p_M(m)\) which models earthquake magnitudes \(m\ge m_0\) independently following an exponential distribution \(p_M(m|\beta )=\beta e^{-\beta (m-m_0)}, \ \beta >0\). Density \(p_M(m)\) corresponds to a Gutenberg-Richter law, and \(m_0\) is the magnitude of completeness, (cut-off magnitude) a threshold above which all events are observed (complete data). . The conditional ETAS intensity function of the ground process can be written as (Ogata 1998)
with \(\varvec{\theta }=(\varvec{\theta }_\mu ,\varvec{\theta }_\varphi )\) a set of parameters. Here the background intensity \(\mu (\varvec{x}|\varvec{\theta }_\mu ): \mathbb {R}^2 \rightarrow [0,\infty )\) defines a non-homogeneous Poisson process in space but stationary in time with \(\varvec{\theta }_\mu \) as the required parameters, while \( \varphi (t-t_i,\varvec{x}-\varvec{x}_i|m_i,\varvec{\theta }_\varphi ):\mathbb {R}^4\rightarrow [0,\infty )\) is the triggering function, modeling the rate of aftershocks (self-exciting process) following an event at \((t_i,\varvec{x}_i)\) with magnitude \(m_i\), controlled by the parameters \(\varvec{\theta }_\varphi \). Specific parametric representations of \(\mu (\cdot )\) and \(\varphi (\cdot )\) for the ETAS model will be discussed in Sect. 2.1.2.
Latent branching structure.
Another interpretation of a Hawkes process (with the ETAS model being a particular example) is as Poisson cluster- or branching process (Hawkes and Oakes 1974), leading to the concept of an underlying branching structure, that is, a non observable latent random variable \(z_i\) for each event i. Events are structured in an ensemble of trees, either having a parent, which is one of the previous events or being spontaneous, called background. The latent variable is typically modelled as taking integer values in a discrete set \(z_i\in \{0,1,\ldots ,i-1\}\), where

Background events defined as \(\mathcal {D}_0=\{(t_i,\varvec{x}_i,m_i,z_i=0)\}_{i=1}^{N_{\mathcal {D}_0}}\) with \(z_i=0\) occur according to a Poisson process with intensity \(\mu (\varvec{x})\) and form cluster centres, i.e., initial points for branching trees. Within each branching tree, an existing event at \(t_j\) can produce direct offspring at \(t>t_j\) according to an inhomogeneous Poisson process with rate \(\lambda _j(t|t_j,\varvec{x}_j,m_j)=\varphi (t-t_j,\varvec{x}-\varvec{x}_j|m_j,\varvec{\theta }_\varphi )\). The overall intensity \(\lambda (t,\varvec{x}|H_t)\) is the sum of all the offspring Poisson processes \(\sum _j \lambda _j\) with \(t_j<t\) and the background Poisson process \(\mu (\varvec{x})\) (Poisson superposition), as given in (1). All events, which are not background, are offspring events defined as \(\mathcal {D}_\varphi =\{(t_i,\varvec{x}_i,m_i,z_i \ne 0)\}_{i=1}^{N_{\mathcal {D}_\varphi }}\), and \(\mathcal {D}=\mathcal {D}_0 \cup \mathcal {D}_\varphi \), where \(\mathcal {D}\) are all the observations.
The latent branching structure cannot be observed. However, by its construction (superposition of i Poisson processes at \(t_i\)) the probability \(p_{i0}=p(z_i=0)\) (background event) is (see,e.g., Zhuang et al. 2002),
while the probability \(p_{ij}=p(z_i=j)\) (event j triggered event i, \(j>0\)) is,
with \(p_{i0}+\sum _j p_{ij}=1\).
2.1.2 Components of the ETAS model
This section sketches the components (background and triggering function) as given in (1).
Background intensity. The background intensity \(\mu (\varvec{x})\) is usually modelled either as piecewise constant function over a rectangular grid (or specific polygones, seismo-tectonic units) with L cells (e.g., in Veen and Schoenberg 2008; Lombardi 2015),
if \(\varvec{x}\) is in grid cell l, \(l=1,\ldots ,L\); or via a weighted kernel density estimator with variable bandwidth, as suggested by Zhuang et al. (2002),
Here, \(|\mathcal {T}|\) is the length of the observational time window, \(p_{i0}\) is the probability that event i is background as defined in (3), \(d_i=\max \{d_{min},r_{i,n_p}\}\) is the variable bandwidth determined for event i corresponding to the distance \(r_{i,n_p}\) of its number of nearest neighbours \(n_p\), where \(d_{min}\) is some minimal bandwidth, and \(k_d(\cdot )\) is an isotropic, bivariate Gaussian kernel function. There are different suggestions to select \(n_p\); Zhuang et al. (2002) propose to choose \(n_p\) between 10 and 100, and state that estimated parameters only change slightly if \(n_p\) is changed in the range of 15–100; Zhuang (2011) suggests based on cross-validation experiments, that an optimal \(n_p\) is in the range 3–6 for Japan. The minimal bandwidth is commonly chosen as \(d_{min}\in [0.02,0.05]\) degrees, which is in the range of the localisation error (Zhuang et al. 2002).
Background parameters to be estimated are \(\varvec{\theta }_\mu =(\mu _1,\mu _2, \ldots ,\mu _L)\) in the first case and the scaled kernel density estimator \(\mu _{\mathrm {kde}}(\varvec{x})\) given through estimated background probabilities \(\{p_{i0}\}_{i=1}^{N_\mathcal {D}}\) in the second case, respectively. For non-parametric models of \(\mu \) as in (6) we neglect the explicit dependency on \(\varvec{\theta }_\mu \) in our notation, but the reader should keep in mind, that in such cases \(\mu \) depends on a varying (potentially infinite) number of parameters.
Parametric triggering function. The triggering function \(\varphi (t-t_i,\varvec{x}-\varvec{x}_i|m_i,\varvec{\theta }_\varphi )\) characterizes the intensity of the offspring processes, thus, the number and distribution of offspring events in space and time. Offspring events (aftershocks) \(\mathcal {D}_\varphi =\{(t_i,\varvec{x}_i,m_i,z_i \ne 0)\}_{i=1}^{N_{\mathcal {D}_\varphi }}\) are triggered by previous events and are all events which are not background. The triggering function \(\varphi (t-t_i,\varvec{x}-\varvec{x}_i|m_i,\varvec{\theta }_\varphi )\) of the ETAS model is usually a non-negative parametric function, which is separable in space and time, and depends on \(m_i\) and \(\varvec{\theta }_\varphi \). There are numerous suggested parameterisations. See, for example, Ogata (1998); Zhuang et al. (2002); Console et al. (2003); Ogata and Zhuang (2006). One of the most common parametrisations is provided by
The first term \(\kappa (\cdot )\) is proportional to the aftershock productivity (or Utsu law, Utsu 1970) of event i with \(m_i\),
and \(K_0\) is called productivity coefficient. Parameters \(K_0\) and \(\alpha \) determine the average number of offspring events (aftershocks) of event i per unit time. The second term \(g(\cdot )\) describes the temporal distribution of aftershocks (offspring); a power law decay proportional to the modified Omori Utsu law (Omori 1894; Utsu 1961), and \(t-t_i>0\) is the elapsed time since the parent event (main shock), that is,
Finally, the third term \(s(\cdot )\) is a probability density function for the spatial distribution of the direct aftershocks (offspring) around the triggering event at \(\varvec{x}_i\). Often, one of the following probability density functions are employed. One distinguishes between a short range decay, which uses an isotropic Gaussian distribution with covariance \(d_1^2 e^{\alpha (m_i-m_0)}\varvec{I}\) (Ogata 1998; Zhuang et al. 2002); and a long range decay following a Pareto distribution (Kagan 2002; Ogata and Zhuang 2006),
where \(\sigma _m(m_i)=d^2 10^{2\gamma (m_i)}\).
The unknown parameters to be estimated are \(\varvec{\theta }_\varphi =(K_0,\alpha ,c,p,d_1)\), or \(\varvec{\theta }_\varphi =(K_0,\alpha ,c,p,d,\gamma ,q)\) depending on which version of \(s(\cdot )\) is used. Note that \(q>1\) and the rest of the parameters are strictly positive.
2.1.3 Parameter estimation via MLE
The likelihood function observing \(\mathcal {D}\) under the spatio-temporal ETAS model is given in (15); it is usually analytically intractable for simple direct optimisation. Numerical optimisation methods (e.g., quasi-Newton methods as in Ogata 1988, 1998, using an EM algorithm Veen and Schoenberg 2008 or simulated annealing Lombardi 2015; Lippiello et al. 2014) are usually employed. Often the integral term related to the triggering function in (15) using (1) is approximated as \(\int _{\mathcal {T}_i}\int _{\mathcal {X}}\sum _{i: t_i< t}\varphi (t-t_i,\varvec{x}-\varvec{x}_i|m_i,\varvec{\theta }_\varphi )\mathop {}\!\mathrm {d}\varvec{x}\mathop {}\!\mathrm {d}t \le \int _{\mathcal {T}_i}\int _{\mathbb {R}^2}\sum _{i: t_i < t}\varphi (t-t_i,\varvec{x}-\varvec{x}_i|m_i,\varvec{\theta }_\varphi )\mathop {}\!\mathrm {d}\varvec{x}\mathop {}\!\mathrm {d}t\), by integrating over \(\mathbb {R}^2\) in space instead of an arbitrary \(\mathcal {X}\) (Schoenberg 2013). The introduced bias is small and often negligible (Schoenberg 2013; Lippiello et al. 2014) while the computations are greatly simplified as \(\int _{\mathbb {R}^2}s(\varvec{x}-\varvec{x}_i|m_i)\mathop {}\!\mathrm {d}\varvec{x}=1\). We also use this approximation. Computational and numerical details of MLE using (15) are given in Ogata (1998). Instead of directly maximising (15), one can augment the likelihood function by a the latent branching structure Z and apply an EM algorithm for MLE (Veen and Schoenberg 2008; Mohler et al. 2011), which is supposed to be advantageous, e.g. regarding stability and convergence (Veen and Schoenberg 2008).
3 Bayesian GP-ETAS model
Our goal is to improve the inference of the spatio-temporal ETAS model in order to allow for comprehensive uncertainty quantification. Despite the availability of powerful MLE based inference methods (see,e.g., Ogata 1998; Veen and Schoenberg 2008; Lippiello et al. 2014; Lombardi 2015), we believe that a Bayesian framework can complement existing methods and will provide a more reliable quantification of uncertainties.
3.1 GP-ETAS model specification
We introduce a novel formulation of the spatio-temporal ETAS model, which models the background rate \(\mu (\varvec{x})\) in a Bayesian non-parametric way via a GP (Williams and Rasmussen 2006), while the triggering function \(\varphi (\cdot )\) assumes still a classical parametric form (modified Omori law (7)). As we will see subsequently, we are able to perform Bayesian inference for this model via Monte Carlo sampling despite its complex form.
While the conditional intensity function of the GP-ETAS model is still given by (1), the background intensity is a priori defined by
where \(\sigma (\cdot )\) is the logistic sigmoid function, \(\bar{\lambda }\) a positive scalar, and \(f(\varvec{x})\) an arbitrary scalar function mapping \(\varvec{x}\in \mathcal {X}\) to the real line \(\mathbb {R}\). Since \(\sigma :\mathbb {R}\rightarrow [0,1]\) the background intensity of the GP-ETAS model is bounded from above by \(\bar{\lambda }\), i.e., \(\mu (\varvec{x})\in [0,\bar{\lambda }]\) for any \(\varvec{x}\in \mathcal {X}\).
For the function \(f(\varvec{x})\) the GP-ETAS model assumes a Gaussian process prior, which implies that the prior over any discrete set of J function values \(\varvec{f}=\{f(\varvec{x}_i)\}_{i=1}^J\) at positions \(\{\varvec{x}_1,\varvec{x}_2,\ldots ,\varvec{x}_J\}\) is a J dimensional Gaussian distribution \(\mathcal {N}(\varvec{f}|\varvec{\mu }_f,\varvec{K}_{\varvec{f},\varvec{f}}),\) where \(\varvec{\mu }_f\) is the prior mean and \(\varvec{K}_{\varvec{f},\varvec{f}}\in \mathbb {R}^{J\times J}\) is the covariance matrix between function values at positions \(\varvec{x}_i\). The matrix \(\varvec{K}_{\varvec{f},\varvec{f}}\) is built from the covariance function (kernel) \(k(\varvec{x},\varvec{x}'|\varvec{\nu })\) such that \(\varvec{K}_{i,j}=k(\varvec{x}_i,\varvec{x}_j|\varvec{\nu })\), where \(\varvec{\nu }\) are hyperparameters. We set \(\varvec{\mu }_f =0\) and employ a Gaussian covariance function
where \( \nu _0\) is the so called amplitude and \((\nu _1,\nu _2)\) are the length scales, representing a distance in input space over which the function values become weakly correlated. Note that the parameter \(\bar{\lambda }\) and the hyperparameters \(\varvec{\nu }\) are also to be inferred from the data. For an in–depth treatment of GPs we refer to Williams and Rasmussen (2006).
The complete specification of the prior model of GP-ETAS including the hyperparameters is now as follows:
The corresponding observational model is

where \(\mathcal {D}\) is the data. Note that some quantities are independent by construction, e.g., \(\varvec{\nu }\) and \(\bar{\lambda }\), f and \(\bar{\lambda }\).
Without the triggering function in the intensity function (1) the GP-ETAS model would be equivalent to the SGCP model which is used to describe an inhomogeneous Poisson process (Adams et al. 2009) because of its favourable statistical properties (Kirichenko and Van Zanten 2015).
In the following we sketch how to generate data from the GP-ETAS model. A full description of the Bayesian inference problem is provided in Sect. 4.
The Figure depicts the different steps of a forward simulation of the generative GP-ETAS model. a Events of a homogeneous Poisson process with intensity \(\bar{\lambda }\) are generated (\(\bar{\lambda }=0.008, N=988\)). b One retains events according to an inhomogeneous Poisson process with the desired intensity \(\mu (\varvec{x})=\bar{\lambda }\sigma (f(\varvec{x}))\) by randomly deleting events (red dots) via thinning. c The background events (black dots from (b)) are denoted by \(\mathcal {D}_0\) (\(N_{\mathcal {D}_0}=481\)), d After adding aftershocks (offspring events) \(\mathcal {D}_\varphi \) to \(\mathcal {D}_0\) in accordance with the triggering function \(\varphi (\cdot )\) one obtains finally the simulated data \(\mathcal {D}\) (\(N_{\mathcal {D}}=2305\)) of the spatio-temporal GP-ETAS model. e Shows the background intensity \(\mu (\varvec{x})=\bar{\lambda }\sigma (f(\varvec{x}))\) together with the generated background events. Gray scaling of the dots refers to the event times. f Depicts the simulated data as a synthetic earthquake catalogue in time
3.2 Simulating the GP-ETAS model
Data \(\mathcal {D}=\{(t_i,\varvec{x}_i,m_i)\}_{i=1}^{N_{\mathcal {D}}}\) can be easily simulated from the GP-ETAS model using the latent branching structure of the point process. We propose a procedure which consists of two parts:
-
1.
Generate all background events \(\mathcal {D}_0=\{(t_i,\varvec{x}_i,m_i,z_i=0)\}_{i=1}^{N_{\mathcal {D}_0}}\) from a SGCP in Eq. (11) as explained in Adams et al. (2009).
-
2.
Sample all aftershock events (offspring) given \(\mathcal {D}_0\) in possibly several generations denoted as \(\mathcal {D}_\varphi =\{(t_i,\varvec{x}_i,m_i,z_i \ne 0)\}_{i=1}^{N_{\mathcal {D}_\varphi }}\) and add them to obtain \(\mathcal {D}=\mathcal {D}_0 \cup \mathcal {D}_\varphi \).
The above procedure can be implemented based on the thinning algorithm (Lewis and Shedler 1976); a variant of rejection sampling for point processes.
After choosing \(\bar{\lambda }\), \(\varvec{\nu }\), \(\varvec{\theta }_\varphi \) and a mark distribution p(m), the simulation procedure of \(\mathcal {D}\in \mathcal {X}\times \mathcal {T}\times \mathcal {M}\) can be summarised as follows: First part: One uses the upper bound \(\bar{\lambda }\) to generate positions \(\{\varvec{x}_j\}_{j=1}^J\) of events from a homogeneous Poisson process with mean \(|\mathcal {X}||\mathcal {T}|\bar{\lambda }\) which provide candidate background events (Fig. 1a). Subsequently a Gaussian process \(\varvec{f}\) is sampled from the prior \(\mathcal {N}(\varvec{f}|\varvec{0},\varvec{K}_{\varvec{f},\varvec{f}})\) based on \(\{\varvec{x}_j\}_{j=1}^J\) using (12). The values \(\mu (\varvec{x}_j)\) can be computed using (11). Afterwards events, which do not follow an inhomogeneous Poisson process with intensity \(\mu (\varvec{x})\) as given by (11), are randomly deleted via thinning (Fig. 1b). The remaining \(N_{\mathcal {D}_0}\) events are background events (Fig. 1c). The event times \(\{t_i\}_{i=1}^{N_{\mathcal {D}_0}}\) are sampled from a uniform distribution \(\mathcal {U}(|\mathcal {T}|)\) and the marks \(\{m_i\}_{i=1}^{N_{\mathcal {D}_0}}\) from an exponential distribution, e.g., Gutenberg-Richter relation. Finally one obtains \(\mathcal {D}_0\). Second part: Given the background events \(\mathcal {D}_0\), the aftershock events (offsprings) of all generations are added to \(\mathcal {D}_0\) in accordance with the triggering function \(\varphi (\cdot )\) and using the mark distribution which yields \(\mathcal {D}\) (Fig. 1d).
The overall simulation algorithm is described in detail in the Appendix 1, and is visualised in Fig. 1.
4 Bayesian inference
In this section, we address the Bayesian inference problem of our spatio-temporal GP-ETAS model. The objective is to estimate the joint posterior density \(p(\mu ,\varvec{\theta }_\varphi | \mathcal {D})\), which encodes the knowledge (including uncertainties) about \(\mu \) and \(\varvec{\theta }_{\varphi }\) after having seen the data. This is because, the posterior density combines information about \(\mu \) and \(\varvec{\theta }_{\varphi }\) contained in the data (via the likelihood function) and prior knowledge (information before seeing the data) about \(\mu \) and \(\varvec{\theta }_{\varphi }\). Here, \(\mu \) denotes the entire random field of the background intensity as in (13d) and \(\varvec{\theta }_{\varphi }\) are the parameters of the triggering function.
The likelihood of observing a point pattern \(\mathcal {D}=\{(t_i,\varvec{x}_i, m_i)\}_{i=1}^{N_{\mathcal {D}}}\) under the GP-ETAS model (11) is given by the point process likelihood
where the intensity \(\lambda (\cdot )\) is given by (11), and the dependencies on \(H_t\), \(H_{t_i}\) are omitted for notational convenience.
Assuming a joint prior distribution denoted here by \(p(\mu ,\varvec{\theta }_\varphi )\) for simplicity, the posterior distribution becomes
This posterior is intractable in practice and hence standard inference techniques are not directly applicable. More precisely, the following three main challenges arise:
-
(i)
The background intensity \(\mu \) and triggering function \(\varphi (\cdot |\varvec{\theta }_\varphi )\) cannot be treated separately in the likelihood function (15).
-
(ii)
The likelihood (15) includes an intractable integral inside the exponential term due to the GP prior on f in (11), that is the integral of f over \(\mathcal {X}\). Furthermore, normalisation of (16) requires an intractable marginalisation over \(\mu \) and \(\varvec{\theta }_\varphi \). Thus, the posterior distribution is doubly intractable (Murray et al. 2006).
-
(iii)
We assume a Gaussian process prior for modelling the background rate. However, the point process likelihood (15) is non-Gaussian, which makes the functional form of the posterior nontrivial to treat in practice.
We approach these challenges by data augmentation based on the work of Hawkes and Oakes (1974), Veen and Schoenberg (2008), Adams et al. (2009), Polson et al. (2013), Donner and Opper (2018). We will find that this augmentation simplifies the inference problem substantially. The following three auxiliary random variables are introduced:
-
(1)
A latent branching structure Z, as described in Sect. 2.1.1, decouples \(\mu \) and \(\varvec{\theta }_\varphi \) in the likelihood function (e.g., Veen and Schoenberg 2008). (See Sect. 4.1 and Eq. (17) for details.)
-
(2)
A latent Poisson process \(\Pi \) enables an unbiased estimation of the integral term in the likelihood function that depends on \(\mu \), as the joint distribution of latent and observed data results in a homogeneous Poisson process with constant integral term. (See Sect. 4.2, paragraph Augmentation by a latent Poisson process and Eq. (21) for details.)
-
(3)
We make use of the fact, that the logistic sigmoid function can be written as an infinite scale mixture of Gaussians using latent Pólya–Gamma random variables \(\omega \sim p_{\scriptscriptstyle \mathrm {PG}}(\omega )\) (Polson et al. 2013), defined in Appendix 2. This leads to a likelihood representation, which is conditional conjugate to all the priors including the Gaussian process prior for the background component of the likelihood function (Donner and Opper 2018). (See Sect. 4.2, paragraph Augmentation by Pólya–Gamma random variables and Eqs. (23, 24) for details.)
These three augmentations allow one to implement a Gibbs sampling procedure (Geman and Geman 1984) that produces samples from the posterior distribution in (16). More precisely, random samples are generated in a Gibbs sampler by drawing one variable (or a block of variables) from the conditional posterior given all the other variables. Hence, we need to derive the required conditional posterior distributions as outlined next.
The suggested sampler consists of three modules using the solutions (data augmentations) sketched above: sampling the latent branching structure, inference of the background \(\mu \), and inference of the triggering \(\varvec{\theta }_\varphi \). Our overall Gibbs sampling algorithm of the posterior distribution is summarised in Algorithm 1. After an initial burn-in (a sufficiently long run of the three modules (Sects. 4.1–4.3), the generated samples converge to the desired joint posterior distribution \(p(\mu ,\varvec{\theta }_\varphi |\mathcal {D})\).

In the following, we discuss some important aspects of the three modules of the Gibbs sampler which the sampler runs repeatedly trough.
4.1 Sampling the latent branching structure
Augmentation by the latent branching structure. We consider an auxiliary variable \(z_i\) for each data point i, which represents the latent branching structure as defined in Sect. 2.1.1. Recall that it gives the time index of the parent event. If \(z_i=0\) then the event is a spontaneous background event. Further we define \(Z=\{z_i\}_{i=1}^{N_{\mathcal {D}}}\), which is the overall branching structure of the data \(\mathcal {D}\). The likelihood \(p(\mathcal {D},Z| \mu ,\varvec{\theta }_\varphi )\) of the augmented model can be written as in 17,
where \({{\,\mathrm{\mathbb {I}}\,}}(\cdot )\) denotes the indicator function, i.e., \({{\,\mathrm{\mathbb {I}}\,}}(z_i=j)\) takes the value 1 for all \(z_i=j\) and 0 otherwise, \(\varphi _{ij}(\varvec{\theta }_\varphi )=\varphi (t_i-t_j,\varvec{x}_i-\varvec{x}_j | m_j,\varvec{\theta }_\varphi )\), \(\varphi _{i}(\varvec{\theta }_\varphi )=\varphi (t-t_i,\varvec{x}-\varvec{x}_i | m_i,\varvec{\theta }_\varphi )\), \(\mathcal {T}_i =[t_i,|\mathcal {T}|] \subset \mathcal {T}\), and all possible branching structures are equally likely, i.e. \(p(Z) = \text{ const }\). Furthermore, \(\mathcal {D}_0=\{\varvec{x}_i\}_{i:z_i=0}\) denotes the set of \(N_{\mathcal {D}_0}\) background events. Note, that marginalizing over Z in (17) recovers (15), because \(\sum _{z_i=0}^{i-1}\mu (\varvec{x}_i)^{{{\,\mathrm{\mathbb {I}}\,}}(z_i=0)}\prod _{j=1}^{i-1} \varphi _{ij}(\varvec{\theta }_\varphi ) ^{{{\,\mathrm{\mathbb {I}}\,}}(z_i=j)}=\lambda (t_i,\varvec{x}_i\vert \mu (\varvec{x}_i),\varvec{\theta }_\phi )\). The augmented likelihood factorises into two independent components, (a) a likelihood component for the background intensity which depends on \(\mu \) (first two terms on the rhs of (17)) and (b) a likelihood component of the triggering function which depends on \(\varvec{\theta }_\varphi \) (last two terms on the rhs of (17)).
From (17) one can derive the conditional distribution of \(z_i\) given all the other variables. Note that all \(z_i\)’s are independent. The conditional distribution is proportional to a categorical distribution,
with the probabilities \(p_{ij}\) given by (3) and (4) which we collect in a vector \(\varvec{p}_i\in \mathbb {R}^i\).
From (18) one can see that the latent branching structure at the kth iteration of the Gibbs sampler is sampled from a categorical distribution, \(\forall i=1,\ldots ,N_{\mathcal {D}}\)
Here \((\mu (\varvec{x}_i),\varvec{\theta }_\varphi )^{(k-1)}\) denotes the values of \(\mu (\varvec{x}_i)\) and \(\varvec{\theta }_\varphi \) from the previous iteration.
4.2 Inference for the background intensity
Given an instance of a branching structure Z, the background intensity in (17) depends on events i for which \(z_i=0\) only. One finds that the resulting term is a Poisson likelihood of the form
where \(\mu (\varvec{x})\) has been replaced by (11) and \(f_i=f(\varvec{x}_i)\) has been used for notational convenience.
Because of the aforementioned problems in Sect. 4, sampling the conditional posterior \(p(f,\bar{\lambda }|\mathcal {D}_0, Z)\) is still non-trivial and requires further augmentations which we describe next.
Augmentation by a latent Poisson process. We can resolve issue (ii) from Sect. 4 by introducing an independent latent Poisson process \(\Pi =\{\varvec{x}_l\}_{l=N_\mathcal {D}+ 1}^{N_{\mathcal {D}\cup \Pi }}\) on the data space with rate \(\hat{\lambda }(\varvec{x})=\bar{\lambda }(1-\sigma (f(\varvec{x})))=\bar{\lambda }(\sigma (-f(\varvec{x})))\) using \(1-\sigma (z)=\sigma (-z)\). The points in \(\mathcal {D}\), \(\Pi \) form the joint set \(\mathcal {D}\cup \Pi \) with cardinality \(N_{\mathcal {D}\cup \Pi }\). Note, that the number of elements in \({\Pi }\), i.e. \(N_{\Pi }\), is also a random variable. The joint likelihood of \(\mathcal {D}_0\) and the new random variable \(\Pi \) is,
where \(f_l=f(\varvec{x}_l)\). Thus, by introducing the latent Poisson process \(\Pi \), we obtain a likelihood representation of the augmented system, where the former intractable integral inside the exponential term disappears, i.e. reduces to a constant.
We can gain some intuition by reminding ourselves of the aforementioned thinning algorithm (Lewis and Shedler 1976) in Sect. 3.2. Considering \(\mathcal {D}_0\) as a resulting set of this algorithm, we wish to find the set \({\Pi }\), such that the joint set \(\mathcal {D}_0\cup {\Pi }\) is coming from a homogeneous Poisson process with rate \(\bar{\lambda }\). Because \(\mathcal {D}_0\) is a sample of a Poisson process with rate \(\bar{\lambda }\sigma (f)\) and the superposition theorem of Poisson processes (Kingman 1993), one finds that if \({\Pi }\) is distributed according to a Poisson process with rate \(\bar{\lambda }\sigma (-f)\), the joint set \(\mathcal {D}_0\cup {\Pi }\) has the rate \(\bar{\lambda }\sigma (f)+\bar{\lambda }\sigma (-f)=\bar{\lambda }\). As we will see later, for the augmented model only the cardinality \(\vert \mathcal {D}_0\cup {\Pi }\vert \) will determine the posterior distribution of \(\bar{\lambda }\).
Having a closer look at the augmented likelihood (21) and considering only terms depending on the function f, one can find a resemblance with a classical classification problem, namely logistic regression. Having the joint set, \(\mathcal {D}_0\cup {\Pi }\) the probability of a point belonging to \(\mathcal {D}_0\) is \(\sigma (f)\) and to \({\Pi }\) it is \(1-\sigma (f)=\sigma (-f)\). Since we know, which points belong to which set, the aim is to find the function f, which best classifies/separates these two sets.
While above we provided some intuition, rigorously one can derive the latent Poisson process \({\Pi }\) following Donner and Opper (2018). Note that (20) implies
where the expectation is over random sets \({\Pi }\) with respect to a Poisson process measure with rate \(\bar{\lambda }\) on the space-time window of the data \(\mathcal {T}\times \mathcal {X}\). Here, one uses Campbell’s theorem (Kingman 1993). Writing the likelihood parts depending on f and \(\bar{\lambda }\) in (17) in terms of the new random variable \({\Pi }\) we get (21). Note that marginalisation over the augmented variable \({\Pi }\) leads back to the background likelihood in (20) conditioned on the branching structure Z.
Note, that at this stage, with the augmentation in this section, our inference problem became tractable, because the augmented likelihood (21) depends on function f only at a finite set of points. In principle at this stage we could employ acceptance rejection algorithms as in Adams et al. (2009). However, to improve efficiency we will introduce one more variable augmentation in the next paragraph, that will allow rejection-free sampling of f.
Augmentation by Pólya–Gamma random variables. Investigating the augmented likelihood (21) issue (iii) from Sect. 4 is still present, because it is nonconjugate to the GP prior that we assume for function f in the GP-ETAS model. However, we noted before, the relation to a logistic regression problem. Polson et al. (2013) introduced the so-called Pólya–Gamma random variables, that allows to efficiently solve the inference problem of logistic GP classifiction (Wenzel et al. 2019). Here, we utilize the same methodology, where make use of the fact that the sigmoid function can be written an infinite scale mixture of Gaussians using latent (Polson et al. 2013), that is,
where the new random Pólya–Gamma variable \(\omega \) is distributed according to the Pólya–Gamma density \(p_{\scriptscriptstyle \mathrm {PG}}(\omega \vert 1,0)\), see Appendix 2. Inserting the Pólya–Gamma representation of the sigmoid function (23) into (21) yields
where we set the Pólya-Gamma variables of all events \(\varvec{\omega }_{\mathcal {D}}=(\omega _1,\ldots ,\omega _{N_\mathcal {D}})\) to \(\omega _i=0\) if \(z_i\ne 0\). For the latent Poisson process the Pólya–Gamma variables are denoted by \(\varvec{\omega }_{{\Pi }}=(\omega _{N_\mathcal {D}+1},\ldots ,\omega _{N_{\mathcal {D}\cup {\Pi }}})\) . The likelihood representation of the augmented system (24) has a Gaussian form with respect to \(\varvec{f}\) (that is, only linear or quadratic terms of \(\varvec{f}\) appear in the exponential function) and is therefore conditionally conjugate to the GP prior denoted by \(p(\varvec{f})\). Hence, we can implement an efficient Gibbs sampler for the background intensity function.
Employing a Gaussian process prior over \(\varvec{f}\) and a Gamma distributed prior over \(\bar{\lambda }\), one gets from (24) the following conditional posteriors for the kth Gibbs iteration:
where \(\varvec{f}=(\varvec{f}_{\mathcal {D}},\varvec{f}_{\Pi }) \in \mathbb {R}^{N_{\mathcal {D}\cup {\Pi }}}\) is the Gaussian process at the data locations \(\mathcal {D}\) and \({\Pi }\); and PP(\(\cdot \)) denotes an inhomogeneous Poisson process with intensity \(\bar{\lambda }(\sigma (-f(\varvec{x})))\); \(\varvec{\Omega }\) is a diagonal matrix with \((\varvec{\omega }_{\mathcal {D}},\varvec{\omega }_{{\Pi }})\) as diagonal entries. \(\varvec{K}\in \mathbb {R}^{N_{\mathcal {D}\cup {\Pi }} \times N_{\mathcal {D}\cup {\Pi }}}\) is the covariance matrix of the Gaussian process prior at positions \(\mathcal {D}\) and \({\Pi }^{(k)}\). It can be shown that, the vector \(\varvec{u}\) is 1/2 for all entries in \(\mathcal {D}_0\), zero for all entries of the remaining data \(\mathcal {D}\backslash \mathcal {D}_0\), and \(-1/2\) for the corresponding entries of \({\Pi }\). Gamma\((\cdot )\) is a Gamma distribution, where the Gamma prior has shape and rate parameters \(\alpha _0,\beta _0\). We used \(e^{-\frac{c^2}{2}\omega } p_{\scriptscriptstyle \mathrm {PG}}(\omega \vert 1,0)\propto p_{\scriptscriptstyle \mathrm {PG}}(\omega \vert 1,c)\) due to the definition of a tilted Pólya–Gamma density (34) as given in (Polson et al. 2013), see Appendix 2. Note that one does not need an explicit form of the Pólya–Gamma density for our inference approach since it is sampling based. In other words, we only need an efficient way to sample from the tilted \(p_{\scriptscriptstyle \mathrm {PG}}\) density (34) which was provided by Windle et al. (2014); Polson et al. (2013). Several \(p_{\scriptscriptstyle \mathrm {PG}}\) samplers are freely available for different computer languages.
In summary, we first introduced a latent Poisson process \({\Pi }\) to render the inference problem of f tractable. The additional Pólya–Gamma augmentation allows us to sample f rejection free, given samples of the augmented sets \({\Pi },\varvec{\omega }_\mathcal {D}, \varvec{\omega }_{\Pi }\). A detailed step-by-step derivation of the conditional distributions is given in the Appendix 3.
Hyperparameters. The Gaussian process covariance kernel given in (12) depends on the hyperparameters \(\varvec{\nu }\). Compare Sect. 3. We use exponentially distributed priors on \(p(\nu _i)=p_{\nu _i}\), and we sample \(\varvec{\nu }\) using a standard MH algorithm as there is no closed form for the conditional posterior available. The only terms where \(\varvec{\nu }\) enter are in the Gaussian process prior and hence the relevant terms are
where \(\varvec{K}_{\varvec{\nu }}\) is the Gaussian process prior covariance matrix depending on \(\varvec{\nu }\) via (12).
4.2.1 Conditional predictive posterior distribution of the background intensity
Given the kth posterior sample \((\bar{\lambda }^{(k)},\varvec{f}^{(k)},\varvec{\nu }^{(k)})\), the background intensity \(\mu (\varvec{x}^*)^{(k)}\) at any set of positions \(\{\varvec{x}_i^*\} \in \mathcal {X}\) (predictive conditional posterior) can be obtained in the following way, see (13d). Conditioned on \(\varvec{f}^{(k)}\) and hyperparameters \(\varvec{\nu }^{(k)}\) the latent function values \(\varvec{f}^*\) can be sampled via the conditional prior \(p(\varvec{f}^*|\varvec{f}^{(k)},\varvec{\nu }^{(k)})\) using (43) with covariance function given in (12) (Williams and Rasmussen 2006). Using (11) one gets \(\mu (\varvec{x}^*)^{(k)}=\bar{\lambda }^{(k)} \sigma (\varvec{f}^*)\).
4.3 Inference for the parameters of the triggering function
Given an instance of a branching structure Z, the likelihood function in (17) factorises in terms involving \(\mu \) and terms involving \(\varvec{\theta }_\varphi \). The relevant terms related to \(\varvec{\theta }_\varphi \) are
The conditional posterior \(p(\varvec{\theta }_\varphi \vert \mathcal {D},Z)\propto p( \mathcal {D}\vert Z, \varvec{\theta }_\varphi )p(\varvec{\theta }_\varphi )\) with prior \(p(\varvec{\theta }_\varphi )\) has no closed form. The dimension of \(\varvec{\theta }_\varphi \) is usually small (\(\le 7\)). We employ MH sampling (Hastings 1970), which can be considered a nested step within the overall Gibbs sampler. We use a random walk MH where proposals are generated by a Gaussian in log space. The acceptance probability of \(\varvec{\theta }_\varphi ^{(k)}\) based on (27) is given by
We take 10 proposals before we return to the overall Gibbs sampler, that is, to step in Sect. 4.1.
5 Experiments and results
We consider two kinds of experiments where we evaluate the performance of our proposed Bayesian approach GP-ETAS (see Sects. 3 and 4). First we look at synthetic data, with known conditional intensity \(\lambda (t,\varvec{x})\), i.e. with known background intensity \(\mu (\varvec{x})\) and known parameters \(\varvec{\theta }_\varphi \) of the triggering function. Here, we investigate if GP-ETAS can recover the model underlying the data well. Secondly, we apply our method to observational earthquake data.
Comparison. We compare our approach with the current standard spatio-temporal ETAS model which uses MLE. This classical ETAS model is based on kernel density estimation with variable bandwidths for the background intensity \(\mu (\varvec{x})\) as described in Sect. 2.1.2. Two variations are considered: (1) ETAS model with standard choice of the minimal bandwidth (0.05 degrees) and \(n_p=15\) the number of nearest neighbors used for obtaining the individual bandwidths (ETAS–classical; Zhuang et al. 2002), and (2) ETAS model with a minimal bandwidth given by Silverman’s rule (Silverman 1986) and \(n_p=15\) (ETAS–Silverman).
Evaluation metrics. Two metrics are used to evaluate the performances. The first metric is the test likelihood, which evaluates the likelihood (15) for a data test set \(\mathcal {D}^*\) (unseen data during the inference) given the inferred model on training data \(\mathcal {D}\), which is \(p(\mathcal {D}^*|\mathcal {D})=\mathbb {E}_{p(\mu ,\varvec{\theta }_\varphi |\mathcal {D})}\left[ p(\mathcal {D}^*|\mu ,\varvec{\theta }_\varphi )\right] \), where the expectation is over the inferred model posterior. The test likelihood reflects the predictive power of the different modelling approaches. In the case of GP-ETAS we obtain K posterior samples \(\{(\mu ^k,\varvec{\theta }^k_{\varphi })\}_{k=1}^K\) and we evaluate the log expected test likelihood, \(\ell _\mathrm{test} = \ln p(\mathcal {D}^*|\mathcal {D}) \approx \ln \frac{1}{K}\sum _{k=1}^K p(\mathcal {D}^*|\mu ^{(k)},\varvec{\theta }^{(k)})\). In the case of ETAS–classical and ETAS–Silverman we use the MLE point estimate for evaluating \(\ell _\mathrm{test}\). The involved spatial integral in (15) is approximated by Riemann sums on a \(50\times 50\) point grid. The second metric is the \(\ell _2\) norm between true background intensity \(\mu \) and the predicted \(\hat{\mu }\), \(\ell _{2} = \sqrt{\int _{\mathcal {X}}(\mu (\varvec{x})-\hat{\mu }(\varvec{x}))^2\mathop {}\!\mathrm {d}\varvec{x}}\). This is only possible for the experiments with synthetic data.
5.1 Synthetic data
General experimental setups. We simulate synthetic data from three different conditional intensity functions which differ in \(\mu (\varvec{x})\). In the first case we consider \(\mu _1(\varvec{x})\) to be constant over large spatial regions, e.g. large area sources (Case 1, Fig. 2 first row). In a second experiment we consider another particular setting where \(\mu _2(\varvec{x})\) is concentrated mainly on small fault-type areas (Case 2, Fig. 2 second row). These two settings (area sources and faults) are important, typical limiting cases in analysing seismicity pattern, both used in seismic hazard assessment. In addition, we consider a third Case (Case 3, Fig. 2 third row), where the transition between high and low background intensity is smooth complementary to Case 1 and Case 2. The chosen intensity functions are,
with \(s=1.5\). The triggering function is given in (7–9) with spatial kernel (10) in all cases. The magnitudes are simulated following an exponential distribution \(p_M(m_i) = \frac{1}{\beta } e^{-\beta (m_i-m_0)}\) with \(\beta =\ln (10)\) which corresponds to a Gutenberg–Richter relation with b-value of 1; \(m_0=3.36\) in the first and third case, \(m_0=3\) in the second case. Spatio-temporal domain is \(\mathcal {X}\times \mathcal {T}=[0,5]\times [0,5]\times [0,5000]\) for Case 1 and Case 3, and \(\mathcal {X}\times \mathcal {T}=[0,5]\times [0,5]\times [0,4000]\) for Case 2. The test likelihood is computed for twelve unseen data sets, simulated from the generative model and averaged. The simulations are done on the same spatial domain \(\mathcal {X}_\mathrm{sim}=[0,5]\times [0,5]\). The time window is \(\mathcal {T}_\mathrm{sim} = [0,1500]\) and \(\ell _\mathrm{test}\) is evaluated using events with event times \(t_i \in [500,1500]\), all previous events are taken into account in the history \(H_t\).
GP-ETAS setup and model robustness. In GP-ETAS we need to set priors on \(\bar{\lambda }\), \(\varvec{\nu }\), \(\varvec{\theta }_{\varphi }\). In addition, we have to choose control parameters of the Gibbs sampler: burn-in B; number of posterior samples K; number of MH steps \(n_\mathrm{MH}\) and width of the proposal distribution \(\sigma _{p_1}\) when sampling the offspring parameters \(\varvec{\theta }_{\varphi }\); and the width of the proposal distribution \(\sigma _{p_2}\) of the MCMC sampling of the hyperparameters \(\varvec{\nu }\). In the following we give some practical guidance on the model setup and we test the robustness of the model using these specifications. A summary of the setup of GP-ETAS is given in Tables 1 and 2.
Hyperparameters \(\varvec{\nu }\) of the covariance function of the GP prior can strongly influence the inference results. We learn \(\varvec{\nu }\) from the data via MCMC sampling in GP-ETAS. We suggest exponentially distributed priors \(p(\nu _i)=\beta _ie^{-\beta _i \nu _i}\) on \(\varvec{\nu }=(\nu _0,\nu _1,\nu _2)\), specified by the prior mean \(\mu _{\nu _i}\). Such a prior choice is common practice for hyperparameter sampling in GP regression. We set \(\mu _{\nu _{i}}=1/\beta _i=c_\nu \mathrm{d}x_i\) for the length scales \(\nu _{1}\) and \(\nu _{2}\) , where \(0.01\le c_\nu \approx 0.1 \le 1\) is a factor, and \(\mathrm{d}x_i=x_{i,\mathrm{max}}-x_{i,\mathrm{min}}\) is the maximum distance in dimension \(x_i\). We choose \(\mu _{\nu _0}=5\) for the amplitude \(\nu _0\). Upper bound \(\bar{\lambda }\) has a Gamma prior, which we specify via its mean \(\mu _{\bar{\lambda }}\) and the uncertainty about this prior mean in terms of the coefficient of variation \(c_{\bar{\lambda }}\). As default set up, we choose \(c_{\bar{\lambda }}=1\) and \(\mu _{\bar{\lambda }}=\frac{ \mathrm{max}({n_1,\ldots ,n_{J}})}{|\mathcal {X}||\mathcal {T}|}\) where \(n_1,\ldots ,n_{J}\) are the number of observations (counts) in spatial bins (2 dimensional histogram count of the positions of the events), and \(N_\mathcal {D}=\sum _{j=1}^{J}n_j\) . We set \(J=100\) bins to cover the whole spatial domain \(\mathcal {X}\) (which corresponds to a 10 by 10 grid). We use uniform priors on the offspring parameters \(\varvec{\theta }_{\varphi }\), given in Table 1. However, expert knowledge of the region of interest can be used to define more appropriate priors on \(\varvec{\nu }\), \(\bar{\lambda }\) and \(\varvec{\theta }_{\varphi }\). Initial values \((\bar{\lambda }, \varvec{\nu }, \varvec{\theta }_{\varphi })^{(0)}\) of the Gibbs sampler can be drawn from the priors. From ML inference of the classical ETAS model it is known that initial values can have an influence on the convergence of the ML optimization and on the obtained results (e.g., Veen and Schoenberg 2008). Unreasonable initial values of \(\varvec{\theta }_{\varphi }\) can prolongate burn-in phase of the GP-ETAS Gibbs sampler. We set following initial values in all our experiments: \(\bar{\lambda }^{(0)}=N_\mathcal {D}/(2|\mathcal {X}||\mathcal {T}|)\) where \(N_\mathcal {D}\) is the number of all observed events and \(|\mathcal {X}|\), \(|\mathcal {T}|\) are the size of the spatial and temporal domain, respectively; \((\nu _{1})^{(0)}\) are initialized using Silverman’s rule based on all observed events; \(\nu _0^{(0)}=5\); \(\varvec{\theta }_{\varphi }^{(0)}=(K_0,c,p,\alpha _m,d,\gamma ,q)^{(0)}=(0.01, 0.01, 1.2, 2.3, \mathrm{d}x_1/100, 0.5, 2.)\) where \(\mathrm{d}x_1=x_{1,\mathrm{max}}-x_{1,\mathrm{min}}\) is the maximum distance in dimension \(x_1\). We choose the control parameters of the Gibbs sampler (see Table 2) based on some pilot simulations. Burn in B is 2000 for Case 1 and Case 2 and \(B=5000\) for Case 3.
In order to test the robustness of our suggested Gaussian process modeling for the background intensity, we perform several synthetic data experiments additionally to those described above. First group of additional experiments: (1) we initialize the Gibbs sampler with a constant \(\mu (\varvec{x})^{(0)}=N_\mathcal {D}/(2|\mathcal {X}||\mathcal {T}|)\) and a very unlikely \(\varvec{\theta }_{\varphi }^{(0)}\) which leads to a sampling of a completely unrealistic branching structure. Here we investigate two cases: (1a) \(\varvec{\theta }_{\varphi }^{(0,a)}\) is set in such a way that only one observation is allocated to the background and all the other events are offspring (aftershocks); (1b) selecting a \(\varvec{\theta }_{\varphi }^{(0,b)}\) in such a manner that all events are considered to be background. Second group of additional experiments: (2) we scale the spatial domain for Case 1 by a factor of 100 in each dimension, i.e. observations are simulated in a spatial domain \(\tilde{\mathcal {X}}=[0,500]\times [0,500]\), temporal domain \(\mathcal {T}\) is kept as before. The generative model is as in Case 1 with two modifications. Background intensity \(\mu _1(\varvec{x})\) is scaled by a factor \(\tilde{c}= |\mathcal {X}|/|\tilde{\mathcal {X}}|\) in order to get the same average number of background events on the scaled spatio-temporal window as in the unscaled case, that is, \(\tilde{\mu }_1(\varvec{x}) =\tilde{c}\mu _1(\varvec{x})\), where \(\tilde{\mu }_1(\varvec{x})\) is the scaled background intensity. In addition, we adjust parameter d of the spatial triggering kernel in (20) in order to obtain a proportional spatial spread of the offspring events, \(\tilde{d} = \tilde{c} d\). We employ the same set up of GP-ETAS as in Case 1 (see Tables 1, 2), in particular we use the same settings regarding the hyperparameters as in Case 1. Additionally, we utilize several initialization of the length scales \(\nu _{1}\), \(\nu _2\) with values ranging from 5 to 500. In both groups of experiments, i.e. (1) and (2), we test if the generative background model can be retrieved and we evaluate the performance measures \(\ell _2\) and \(\ell _\mathrm{test}\).
Experimental results of background intensity \(\mu _1(\varvec{x})\) for the synthetic data of Case 1: First and second row: a generative model, b median GP-ETAS, c uncertainty GP-ETAS as semi inter quantile 0.05, 0.95 distance d ETAS–classical MLE, e ETAS–Silverman MLE, f normalised histogram of the sampled upper bound \(\bar{\lambda }\). Dots are the background events of the realisation. Third and fourth row: One dimensional profiles of \(\mu _1(\varvec{x})\) (ground truth) and inferred results are shown. The profiles are at \(y\in \{0.5,2,3,4.5\}\) and \(x\in \{1,2,2.5,3.5\}\)
Results of these additional experiments related to the robustness of GP-ETAS are shown in Appendix 4. The model of the background intensity underlying the data can be recovered in an acceptable way, in a sensible length of iterations in all experiments. GP-ETAS performs better than standard ETAS models for both metrics \(\ell _2\) and \(\ell _\mathrm{test}\). Beyond the investigated cases there might be scenarios in which the model fails, but under reasonable assumptions (from application point of view) regarding priors, initial values, control parameters of the sampler and based on a sufficient amount of data, our proposed GP-ETAS seems to be robust.
Findings and interpretations. The ground truth and inferred results of \(\mu (\varvec{x})\) and \(\varvec{\theta }_\varphi \) are given in Figs. 3, 4 and 5 and Tables 3, 4 and 5. Performance metrics: the averaged \(\ell _\mathrm{test}\) of twelve unseen data sets and the numerically approximated error of the estimated background intensity \(\ell _2\) are shown in Table 6. Here we describe a few noteworthy aspects. First of all, GP-ETAS recovers well the assumptions both the background intensity \(\mu (\varvec{x})\) and the parameters of the triggering function \(\varvec{\theta }_\varphi \). GP-ETAS outperforms the standards models for both metrics \(\ell _\mathrm{test}\) and \(\ell _2\). The latter fact is of particular importance, as it is common practice to use the declustered background intensity \(\mu (\varvec{x})\) for seismic hazard assessment. One may appreciate that ETAS–classical occasionally tends to strongly overshoot the true \(\mu (\varvec{x})\) (see Fig. 3); in regions with many aftershocks, e.g. near (2.8,4.5), (1.2,3.8). This effect is less pronounced for ETAS–Silverman, where the minimum bandwidth is broader. In our approach no bandwidth selection has to be made in advance, it is obtained via sampling the hyperparameters. One also observes, that ETAS–classical and ETAS–Silverman suffer more strongly from edge effects than GP-ETAS, which seems to be fairly unaffected.
The parameters of the triggering function \(\varvec{\theta }_\varphi \) are roughly correctly identified in all cases and methods. All the values are close to those of the generative model. All the methods overestimate c and \(K_0\) (in Case 2), however the true values are still included in the uncertainty band (credible band) of GP-ETAS. GP-ETAS has the advantage that it provides the whole distribution of the parameters instead of only a point estimate. The median of the obtained upper bound \(\bar{\lambda }\) on \(\mu (\varvec{x})\) using GP-ETAS overestimates in Case 1 (underestimates in Case 2) the true upper bound, however, it is fairly close to the true value, which is contained in the uncertainty band around \(\bar{\lambda }\). The median of \(\bar{\lambda }\) is substantially underestimated in Case 3, here it is not contained in the uncertainty band.
Computational costs. GP-ETAS has approximately a complexity of \(\mathcal {O}((N_{\mathcal {D}\cup {\Pi }})^3)\) for the inference of \(\mu \). This is due to the matrix inversions involved in Gaussian process modelling. The estimation of \(\varvec{\theta }_\varphi \) is less expensive and approximately of \(\mathcal {O}(N_\mathcal {D}^2)\), where \(N_\mathcal {D}\) is the number of data points. In addition, the number of required samples in order to obtain a valuable approximation of the posterior distribution depends on the mixing properties of the Markov chain. From our experience based on the performed experiments one needs \(>10^3\) samples after a burn-in phase of \(>10^3\) iterations. Therefore, our proposed method in its current implementation is computationally expensive but still feasible for small to intermediate data sets with approximately \(N_\mathcal {D}\lessapprox 10^4\) events; which seems sufficient for many situations where site specific seismic analysis takes place. Run times of GP-ETAS model are given in Appendix 5.
5.2 Case study: L’Aquila, Italy
Now we apply GP-ETAS to real data and compare the performance with the other models ETAS–classical and ETAS–Silverman.
The L’Aquila region in central Italy is seismically active and experiences from time to time severe earthquakes. The most famous example is the \(M_w=6.2\) earthquake on 6 April 2009, which occurred directly below the City of L’Aquila, and caused large damage and more than 300 deaths (Marzocchi et al. 2014). This event was followed by a seismic sequence with a largest earthquake of \(M_w=4.2\), latter occurred almost one year later on 30 March 2010 (Marzocchi et al. 2014).
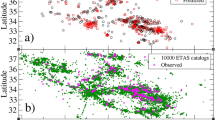
The L’Aquila data set comprises \(N=2189\) events which occurred in a time period from 04/02/2001 to the 28/3/2020, on a spatial domain \(\mathcal {X}=[12^{\circ } E,15^{\circ } E]\times [41^{\circ } N,44^{\circ }N]\) with earthquake magnitudes \(3.0\le m \le 6.5\). The data was obtained from the website of the National Institute of Geophysics and Vulcanology of Italy (http://terremoti.ingv.it/, Istituto Nazionale della Geofisica e Vulcanologia, INGV). We split the data set into training data, all events with event times \(t_i\le 4000\) days (\(N_\mathrm{training}=723\) events, \(\mathcal {T}_\mathrm{training}=[0,4000]\) days), and test data, all events with \(t_i>4000\) days (\(N_\mathrm{test}=1466\), \(\mathcal {T}_\mathrm{test}=[4000,6993]\) days), as shown in Fig. 6. The training data is used for the inference and the test data is used to evaluate the performance of the different models.
The inference setup is the same as described for the synthetic data. We simulate 15,000 posterior samples after a burn in of 2000. The priors are—as for the synthetic data—given in Tables 1 and 2 . The inference results for \(\mu (\varvec{x})\) and \(\varvec{\theta }_\varphi \) are shown in Fig. 7 and Table 7. The performance metric \(\ell _\mathrm{test}\) for different unseen data sets (next 30 days, one year in future, five years in future, and the total test data \(\approx 8\) years) is shown in Table 8. The posterior distribution of the upper bound of the background intensity is shown in Fig. 8.
For the l’Aquila data set GP-ETAS performs slightly better than the other models in terms of \(\ell _\mathrm{test}\). Note, that ETAS–classical estimates fairly large values for \(\mu (\varvec{x})\) in regions with many aftershocks (Fig. 7). This is similar to Case 1 for the synthetic experiments. Hence, as for the synthetics one may assume that ETAS–classical overshoots in these regions; the posterior of \(\bar{\lambda }\) supports this hypothesis, see Fig. 8. The estimated \(\varvec{\theta }_\varphi \) are similar for all models. Posterior samples and MLE estimates of d, \(\gamma \), q are shown in Fig. 9. The spatial kernels of the three models are shown in Fig. 10 for the mean magnitude and a large magnitude.
Results real data, L’Aquila data set: background intensity \(\mu (\varvec{x})\) [number of shocks with \(m \ge 3\) /day/degree\(^2\)] a ETAS–classical MLE, b ETAS–Silverman MLE, c median GP-ETAS, d uncertainty GP-ETAS: semi inter quantile 0.05, 0.95 distance, and dots are the events of the training data, where the grey scaling depicts the event times, from black (older events) to white (current events). Note, a–c have the same scale
6 Discussion and conclusions
We have demonstrated that the proposed GP-ETAS model allows for augmentation techniques (Hawkes and Oakes 1974; Veen and Schoenberg 2008; Adams et al. 2009; Donner and Opper 2018) and hence provides the means of assessing the Bayesian posterior of a semi-parametric spatio-temporal ETAS model. We have shown for three examples that the predictive performance improves over classical methods. In addition, we can quantify parameter uncertainties via their empirical posterior density. The developed framework is flexible and allows for several extensions that deserve consideration in future research, e.g. a time depending on background rate. Another obvious extension of our work would be a Bayesian non-parametric treatment of the triggering function \(\varphi \). See also (Zhang et al. 2019b).
As mentioned earlier Kolev and Ross (2020) propose an alternative semiparametric framework of the ETAS model, which assumes a Dirichlet process prior over the background intensity. Such a model is more related to the classical KDE approach than the model proposed in our work. It considers, that the background intensity is modelled via density over scaled mixture models. We expect, that inference of the Dirichlet process will be less costly than the for the GP-ETAS model. In contrast, to define the GP-prior seems more intuitive, because one mainly needs to impose some spatial correlation structure, while for the Dirichlet prior we need to have an idea, how many components model well the background intensity function on average. However, which of the two models will be the more suitable one, might depend on the specific data instance.
Future research will deal with a more geology informed choice of the prior GP. Sometimes a given catalog comes with information about, e.g. fault locations, which are not straightforward to incorporate in traditional treatments of the spatio-temporal ETAS model. For the GP-ETAS model, however, incorporation of informative priors is possible within our Bayesian setting. For example, spatial information about fault zones can be incorporated by an adequate choice of the mean of the GP, which was chosen to be 0 throughout this work. While we restricted ourselves to the squared exponential (12) as covariance function for the GP prior of f the framework is not restricted to this either and other covariance function can be used to incorporate prior information, e.g. from the Matérn class, or any other function that ensures that the covariance matrix is positive definite.
Another important issue is the computational effort. Having to sample the GP at all observed events in \(\mathcal {D}\) and at positions of the latent Poisson process \({\Pi }\), resulting in a cubic complexity of \(\mathcal {O}\left( (N_{\mathcal {D}\cup {\Pi }})^3\right) \), implies an undesirable computational complexity of the current GP-ETAS Gibbs sampler. There are, however, several possibilities to mitigate this complexity via model approximations and/or alteration. For example, one could resort to approximations to the posterior distribution in order to be able to scale the GP-ETAS model to larger catalogues (\(N_D\gg 10^3\)). While those always come with the sacrifice of asymptotic exactness, some approaches are likely to provide good estimates in the large data regime. One of such approximations is provided by variational inference, which was already proposed for the SGCP by Donner and Opper (2018) utilising sparse GPs (Titsias 2009). This approach makes use of the same model augmentations utilised in this work. The variational posterior of the triggering parameters could be inferred, e.g., via black-box variational inference (Ranganath et al. 2014; Gianniotis et al. 2015). Alternatively, one could restrict the calculations to finding the MAP estimate of the GP-ETAS model. For the background intensity this can be efficiently done by an expectation-maximisation algorithm based on the model augmentations presented here and sparse GPs (Donner and Opper 2018). This can be combined with a Laplace approximation to provide an approximate Gaussian posterior. The limiting factor under such approximations will most likely arise from the branching structure for which the required computations scale like \(\mathcal{O}(N_\mathcal {D}(N_\mathcal {D}- 1) / 2)\). Finally, one could also investigate gradient-free affine invariant sampling methods, as proposed by Reich and Weissmann (2019); Garbuno-Inigo et al. (2020, 2019).
We conclude by re-emphasising the importance of semi-parametric Bayesian approaches to spatio-temporal statistical earthquake modelling and the need for developing efficient tools for their computational inference. Within this work, we combined the SGCP model (Adams et al. 2009) for the background intensity with the ETAS model, such that a Gibbs sampling approach could be made tractable and efficient via specific data augmentation. Finally, we have demonstrated the model’s applicability to realistic earthquake catalogs.
References
Adams, R.P., Murray, I., MacKay, D.J.: Tractable nonparametric Bayesian inference in Poisson processes with Gaussian process intensities. In: Proceedings of the 26th Annual International Conference on Machine Learning, pp 9–16 (2009)
Adelfio, G., Chiodi, M.: Alternated estimation in semi-parametric space-time branching-type point processes with application to seismic catalogs. Stoch. Environ. Res. Risk Assess. 29(2), 443–450 (2014)
Bacry, E., Muzy, J.F.: First- and second-order statistics characterization of Hawkes processes and non-parametric estimation. IEEE Trans. Inf. Theory 62, 2184–2202 (2016)
Bacry, E., Mastromatteo, I., Muzy, J.F.: Hawkes processes in finance. Mark. Microstruct. Liq. 1(01), 1550005 (2015)
Console, R., Murru, M., Lombardi, A.M.: Refining earthquake clustering models. J. Geophys. Res. Solid Earth 108(B10), 1–9 (2003)
Daley, D., Vere-Jones, D.: An Introduction to the Theory of Point Processes. Vol. I: Elementary Theory and Methods, 2nd edn. Springer, New York (2003)
Dempster, A.P., Laird, N.M., Rubin, D.B.: Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B (Methodol.) 39(1), 1–22 (1977)
Donner, C., Opper, M.: Efficient Bayesian inference of sigmoidal Gaussian cox processes. J. Mach. Learn. Res. 19(1998), 1–34 (2018)
Donnet, S., Rivoirard, V., Rousseau, J.: Nonparametric Bayesian estimation of multivariate Hawkes processes. arXiv preprint arXiv:180205975 (2018)
Filimonov, V., Sornette, D.: Apparent criticality and calibration issues in the Hawkes self-excited point process model: application to high-frequency financial data. Quant. Finance 15(8), 1293–1314 (2015)
Fox, E.W., Schoenberg, F.P., Gordon, J.S.: Spatially inhomogeneous background rate estimators and uncertainty quantification for nonparametric Hawkes point process models of earthquake occurrences. Ann. Appl. Stat. 10(3), 1725–1756 (2016)
Garbuno-Inigo, A., Nüsken, N., Reich, S.: Affine invariant interacting Langevin dynamics for Bayesian inference. Technical report. arXiv:1912.02859, SIAM J. Dyn. Syst. (in press) (2019)
Garbuno-Inigo, A., Hoffmann, F., Li, W., Stuart, A.: Interacting Langevin diffusions: gradient structure and ensemble Kalman sampler. SIAM J. Appl. Dyn. Syst. 19, 412–441 (2020)
Geman, S., Geman, D.: Stochastic relaxation, Gibbs distributions, and the Bayesian restoration of images. IEEE Trans. Pattern Anal. Mach. Intell. PAMI 6(6), 721–741 (1984)
Gerhard, F., Deger, M., Truccolo, W.: On the stability and dynamics of stochastic spiking neuron models: nonlinear Hawkes process and point process GLMs. PLoS Comput. Biol. 13(2), 1–31 (2017)
Gianniotis, N., Schnörr, C., Molkenthin, C., Bora, S.S.: Approximate variational inference based on a finite sample of Gaussian latent variables. Pattern Anal. Appl. 19(2), 475–485 (2015)
Hastings, W.K.: Monte Carlo sampling methods using Markov chains and their applications. Biometrika 57(1), 97–109 (1970)
Hawkes, A.G.: Spectra of some self-exciting and mutually exciting point processes. Biometrika 58(1), 83–90 (1971)
Hawkes, A.G., Oakes, D.: A cluster process representation of a self-exciting process. J. Appl. Probab. 11(3), 493–503 (1974)
Hu, W., Jin, P.J.: An adaptive Hawkes process formulation for estimating time-of-day zonal trip arrivals with location-based social networking check-in data. Transp. Res. Part C Emerg. Technol. 79, 136–155 (2017)
Jordan, T.H., Chen, Y.T., Gasparini, P., Madariaga, R., Main, I., Marzocchi, W., Papadopoulos, G., Sobolev, G., Yamaoka, K., Zschau, J.: Operational earthquake forecasting. State of knowledge and guidelines for utilization. Ann. Geophys. 54(4), 315–391 (2011)
Kagan, Y.Y.: Aftershock zone scaling. Bull. Seismol. Soc. Am. 92(2), 641–655 (2002)
Kingman, J.F.C.: Poisson Processes. Oxford University Press, Oxford (1993)
Kirchner, M.: An estimation procedure for the Hawkes process. Quant. Finance 17(4), 571–595 (2017)
Kirichenko, A., Van Zanten, H.: Optimality of Poisson processes intensity learning with Gaussian processes. J. Mach. Learn. Res. 16(1), 2909–2919 (2015)
Kolev, A.A., Ross, G.J.: Semiparametric Bayesian forecasting of spatial earthquake occurrences. arXiv preprint arXiv:200201706 (2020)
Lewis, P.A., Shedler, G.S.: Simulation of nonhomogeneous Poisson processes with log linear rate function. Biometrika 63(3), 501–505 (1976)
Linderman, S.W., Adams, R.P.: Scalable Bayesian inference for excitatory point process networks. arXiv preprint arXiv:150703228 (2015)
Lippiello, E., Giacco, F., de Arcangelis, L., Marzocchi, W., Godano, C.: Parameter estimation in the ETAS model: approximations and novel methods. Bull. Seismol. Soc. Am. 104(2), 985–994 (2014)
Lombardi, A.M.: Estimation of the parameters of ETAS models by simulated annealing. Sci. Rep. 5(8417), 1–11 (2015)
Marsan, D., Lengliné, O.: Extending earthquakes’ reach through cascading. Science 319(5866), 1076–1079 (2008)
Marzocchi, W., Lombardi, A.M., Casarotti, E.: The establishment of an operational earthquake forecasting system in Italy. Seismol. Res. Lett. 85(5), 961–969 (2014)
Mohler, G.O., Short, M.B., Brantingham, P.J., Schoenberg, F.P., Tita, G.E.: Self-exciting point process modeling of crime. J. Am. Stat. Assoc. 106(493), 100–108 (2011)
Murray, I., Ghahramani, Z., MacKay, D.J.: MCMC for doubly-intractable distributions. In: Proceedings of the 22nd Conference on Uncertainty in Artificial Intelligence, UAI 2006, pp. 359–366 (2006)
Ogata, Y.: Asymptotic behavoir of maximum likelihood. Ann. Inst. Stat. Math. 30, 243–261 (1978)
Ogata, Y.: Statistical models for earthquake occurrences and residual analysis for point processes. J. Am. Stat. Assoc. 83, 9–27 (1988)
Ogata, Y.: Space-time point-process models for earthquake occurrences. Ann. Inst. Stat. Math. 50(2), 379–402 (1998)
Ogata, Y., Zhuang, J.: Space-time ETAS models and an improved extension. Tectonophysics 413(1–2), 13–23 (2006)
Omori, F.: On the after-shocks of earthquakes. J. Coll. Sci. 7, 111–120 (1894)
Polson, N.G., Scott, J.G., Windle, J.: Bayesian inference for logistic models using Pólya–Gamma latent variables. J. Am. Stat. Assoc. 108(504), 1339–1349 (2013)
Porter, M.D., White, G.: Self-exciting hurdle models for terrorist activity. Ann. Appl. Stat. 4(1), 106–124 (2010)
Ranganath, R., Gerrish, S., Blei, D.M.: Black box variational inference. In: Proceedings of the Seventeenth International Conference on Artificial Intelligence and Statistics (2014)
Rasmussen, J.G.: Bayesian inference for Hawkes processes. Methodol. Comput. Appl. Probab. 15(3), 623–642 (2013)
Rathbun, S.L.: Asymptotic properties of the maximum likelihood estimator for spatio-temporal point processes. J. Stat. Plan. Inference 51(1), 55–74 (1996)
Reich, S., Weissmann, S.: Fokker–Planck particle systems for Bayesian inference: computational approaches. Technical report. arXiv:1911.10832, University of Potsdam (2019)
Reynaud-Bouret, P., Schbath, S.: Adaptive estimation for Hawkes processes; application to genome analysis. Ann. Stat. 38(5), 2781–2822 (2010)
Ross, G.: Bayesian estimation of the ETAS model for earthquake occurrences. Preprint (2018)
Schoenberg, F.P.: Facilitated estimation of ETAS. Bull. Seismol. Soc. Am. 103(1), 601–605 (2013)
Silverman, B.W.: Density Estimation for Statistics and Data Analysis, vol. 26. CRC Press, Boca Raton (1986)
Titsias, M: Variational learning of inducing variables in sparse Gaussian processes. In: Artificial Intelligence and Statistics, pp 567–574 (2009)
Utsu, T.: A statistical study on the occurrence of aftershocks. Geophys. Mag. 30, 521–605 (1961)
Utsu, T.: Aftershocks and earthquake statistics (1): some parameters which characterize an aftershock sequence and their interrelations. J. Fac. Sci. Hokkaido Univ. Ser. 7 Geophy. 3(3), 129–195 (1970)
Veen, A., Schoenberg, F.P.: Estimation of space-time branching process models in seismology using an EM-type algorithm. J. Am. Stat. Assoc. 103(482), 614–624 (2008)
Wang, Q., Schoenberg, F.P., Jackson, D.D.: Standard errors of parameter estimates in the ETAS model. Bull. Seismol. Soc. Am. 100(5 A), 1989–2001 (2010)
Wenzel, F., Galy-Fajou, T., Donner, C., Kloft, M., Opper, M.: Efficient gaussian process classification using Pòlya–Gamma data augmentation. Proc. AAAI Conf. Artif. Intell. 33, 5417–5424 (2019)
Williams, C.K., Rasmussen, C.E.: Gaussian Processes for Machine Learning. MIT Press, Cambridge, MA (2006)
Windle, J., Polson, N.G., Scott, J.G.: Sampling Pólya–Gamma random variates: alternate and approximate techniques. arXiv preprint arXiv:14050506 (2014)
Zhang, R., Walder, C., Rizoiu, M.A.: Variational inference for sparse Gaussian process modulated Hawkes process. arXiv preprint arXiv:1905.10496 (2019a)
Zhang, R., Walder, C., Rizoiu, M.A., Xie, L.: Efficient non-parametric Bayesian Hawkes processes. In: Proceedings of the Twenty-Eighth International Joint Conference on Artificial Intelligence, IJCAI-19, pp. 4299–4305 (2019b)
Zhao, Q., Erdogdu, M.A., He, H.Y., Rajaraman, A., Leskovec, J.: SEISMIC: a self-exciting point process model for predicting tweet popularity. In: Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp 1513–1522 (2015)
Zhou, K., Zha, H., Song, L.: Learning social infectivity in sparse low-rank networks using multi-dimensional Hawkes processes. In: Proceedings of the International Conference on Artificial Intelligence and Statistics, pp. 641–649 (2013)
Zhou, F., Li, Z., Fan, X., Wang, Y., Sowmya, A., Chen, F.: Scalable inference for nonparametric Hawkes process using Pólya–Gamma augmentation. arXiv:1910.13052v1 (2019)
Zhuang, J.: Next-day earthquake forecasts for the Japan region generated by the ETAS model. Earth Planets Space 63(3), 207–216 (2011)
Zhuang, J., Ogata, Y., Vere-Jones, D.: Stochastic declustering of space-time earthquake occurrences. J. Am. Stat. Assoc. 97(458), 369–380 (2002)
Acknowledgements
This research has been partially funded by Deutsche Forschungsgemeinschaft (DFG, German Science Foundation) - SFB 1294/1 - 318763901.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendices
Appendices
1.1 A GP-ETAS generative model
The generative model of GP-ETAS consists of two parts given in Algorithm 2–3, and requires several inputs. Here, we chose a GP with zero mean and covariance function \(k(\varvec{x},\varvec{x}\prime \vert \varvec{\nu })\) given in (12) with hyperparameters \(\varvec{\nu }\), a triggering function \(\varphi (\cdot |\varvec{\theta }_\varphi )\) given in (7–9) with spatial kernel (10) and therefore \(\varvec{\theta }_\varphi =(K_0,c,p,\alpha ,d,\gamma ,q)\), and a mark distribution, that is an exponential distribution \(m_i-m_0 \sim \mathrm{Exponential}(\beta )\) (Gutenberg–Richter relation) with parameters \(\beta ,m_0\). The simulation algorithm can be easily adjusted for other choices.


1.2 B Definition of the Pólya–Gamma density
Here, we briefly define the Pólya–Gamma density (Polson et al. 2013). First we define the \(p_{\scriptscriptstyle \mathrm {PG}}(\omega \vert b,0)\), which is completely defined through its Laplace transform
With this definition it can be shown that
where \(g_k\sim \mathrm {Gamma}(b,1)\) and the equality is in distribution. With this result one can then define a tilted Pólya–Gamma density given by
where \(b\in \mathbb {R}^+\) and \(c\in \mathbb {R}\), and the normalisation can be straightforwardly obtained with (32). Also for this the tilted density we can derive the Laplace transform
From (35) all moments of the Pólya–Gamma density can be derived analytically and furthermore an acceptance-rejection algorithm with high acceptance rate was derived by Polson et al. (2013).
1.3 C Conditional posteriors for the background intensity
Here we derive the conditional posterior distributions given in (25a)–(25e). For each of those our starting point is the augmented likelihood of the background intensity in (24) with the prior of interest. Note, that for the augmented variables \({\Pi },\varvec{\omega }_{\mathcal {D}},\varvec{\omega }_{\Pi }\) there are no additional priors, and hence their conditionally distribution will only be determined by (24).
The latent Poisson process \({\Pi }\). To derive the conditional posterior for \({\Pi }\), we consider all terms in (24) that depend on \({\Pi }=\{x_l\}_{l=1+N_\mathcal {D}}^{N_{\mathcal {D}\cup {\Pi }^{(k)}}}\) and marginalise over the \(\varvec{\omega }_{{\Pi }}\). This results in
which we identify as an unnormalised Poisson process density with rate \(\bar{\lambda }\sigma (-f_l)\). Again note, that the process is defined over \(\mathcal {T}\times \mathcal {X}\). To sample \({\Pi }^{(k)}\) in the \(k^\mathrm{th}\) iteration, we can utilise the thinning procedure (Lewis and Shedler 1976), where we first sample a homogeneous point process with intensity \(\bar{\lambda }^{(k-1)}\). At the resulting points we draw the the GP f given the previous sample \(\varvec{f}^{(k-1)}\) from the predictive distribution (see below). Then, we keep all events l with probability \(\sigma (-f_l)\) which yields \({\Pi }^{(k)}\) according to (25a).
The Pólya–Gamma random variables \(\varvec{\omega }\). Next we sample the Pólya–Gamma random variables at \(\mathcal {D}_0\). From (24) we see, that the components in \(\varvec{\omega }_{\mathcal {D}_0}\) factorise, meaning that the conditional posteriors are independent. Hence, we get for each \(i:z_i=0\)
and hence \(\varvec{\omega }_{\mathcal {D}}^{(k)}\) can be sampled independently from a tilted Pólya–Gamma distribution where \(i:z_i=0\), given branching structure \(Z^{(k)}\) and GP \(\varvec{f}^{(k-1)}\). For \(i:z_i\ne 0\) we set \(\omega _i=0\). The last equivalence follows from a property of an tilted Pólya–Gamma distribution, see (34). In effect, we get (25c).
The conditional posterior \(\varvec{\omega }_{{\Pi }}\) at the positions of the latent events \({\Pi }\) also factorises in all components and hence we get for each \(l=N_\mathcal {D}+ 1, \ldots ,N_{\mathcal {D}\cup {\Pi }}\)
and also \(\varvec{\omega }_{{\Pi }}^{(k)}\) can be sampled independently from a tilted Pólya–Gamma distribution (25b) given the samples of \({\Pi }^{(k)}\) and \(\varvec{f}^{(k-1)}\); we get (25b).
The upper bound on the intensity \(\bar{\lambda }\). For \(\bar{\lambda }\) we assume a Gamma prior \(p(\bar{\lambda }\vert \alpha _0,\beta _0)\) with shape parameter \(\alpha _0\) and rate parameter \(\beta _0\). Together with (24) we derive the conditional posterior being,
where \(\alpha _1 = N_{\mathcal {D}_0\cup {\Pi }}+\alpha _0\) and \(\beta _1 = |\mathcal {T}||\mathcal {X}|+\beta _0\). Hence, given \({\Pi }^{(k)}\) and \(D_0^{(k)}\) we can sample \(\bar{\lambda }^{(k)}\), and one gets (25d).
The posterior Gaussian Process f. For the conditional posterior of the Gaussian process f we rewrite (24) with the terms depending on f as follows
where we define \(u_i=\frac{1}{2}\) if \(z_i=0\), \(u_i=0\) if \(z_i\ne 0\), and \(u_l=-\frac{1}{2}\) for \(l=N_\mathcal {D}+1,\ldots ,N_{\mathcal {D}\cup {\Pi }}\). It follows that \(\varvec{\Omega }=\mathrm{diag}(\varvec{\omega }_\mathcal {D},\varvec{\omega }_{{\Pi }})\). The GP prior \(\varvec{f}\) at a finite set \(\mathcal {D},{\Pi }\) of points is given by
where the entry of row i and column j of \(\varvec{K}_{\varvec{f},\varvec{f}}\) is given by the covariance function (12) \(k(\varvec{x}_i, \varvec{x}_j \vert \varvec{\nu })\). Together with (40) we identify the conditional posterior
which defines the conditional posterior at \(\varvec{f}\), and it is easy to sample \(\varvec{f}^{(k)}\) given instances of \(\varvec{\omega }_{\mathcal {D}}, {\Pi }, \varvec{\omega }_{{\Pi }}\) from previous samples. However, the algorithm requires us to sample the posterior also at other points \(\varvec{x}^*\notin \mathcal {D}\cup {\Pi }\), e.g. for sampling the next instance of \({\Pi }\) or for visualisation of the background intensity \(\mu (\varvec{x})\) on a grid. Here, we denote the GP at all additional points \(\varvec{f}^*\). The GP prior defines a predictive distribution (Williams and Rasmussen 2006) of any set \(\varvec{f}^*\) given \(\varvec{f}\)
where \(\varvec{K}_{\varvec{f}^*,\varvec{f}^*}\) contains the covariances between the points \(\varvec{x}^*\) of \(\varvec{f}^*\) and \(\varvec{K}_{\varvec{f}^*,\varvec{f}}=\varvec{K}_{\varvec{f}^*,\varvec{f}}^\top \) between the points \(\varvec{x}^*\) of \(\varvec{f}^*\) and \(\varvec{f}\) at \(\varvec{x}\). Note, that the posterior of \(\varvec{f}^*\) given \(\varvec{f}\) is equal to the conditional prior, since (24) does not depend on \(\varvec{f}^*\).
1.4 D Additional experiments
Here, we report a summary of the results related to additional synthetic data experiments described in paragraph ’GP-ETAS set up and model robustness’ of Sect. 5.1 in the main text.
Results of experiment 1. We investigate two experiments: (1a) an initialization of the Gibbs sampler with almost only offspring events and just one background event; (1b) vice versa an initialization of the Gibbs sampler with almost only background events and no or only a few offspring events. In order to obtain such extreme realizations of the branching structure (1a and 1b), we choose initial offspring parameters \(\varvec{\theta }_{\varphi }^{(0)}=(K_0,c,p,\alpha _m,d,\gamma ,q)\) as follows: Experiment (1a) \(\varvec{\theta }_{\varphi }^{(0,\mathrm{a})}=(10., 0.01, 1.2, 10., 0.05, 0.5, 2.)\); Experiment (1b) \(\varvec{\theta }_{\varphi }^{(0,\mathrm{b})}=(1\mathrm{e}-06, 0.01, 1.2, 2.3, 0.05, 0.5, 2.)\); initialization of the background intensity is \(\mu (\varvec{x})^{(0)}=N_{\mathcal {D}}/(2|\mathcal {X}||\mathcal {T}|)\) in both experiments. Underlying generative models are those of Case 1, Case 2 and Case 3.
The Gibbs sampler converges after a sensible number of iterations (\(<2000\)) to the ground truth in terms of the number of events assigned to the background process \(\mu (\varvec{x})\), as depicted in Fig. 11. The generative models, i.e. background intensities \(\mu _1(\varvec{x})\), \(\mu _2(\varvec{x})\) and \(\mu _3(\varvec{x})\) together with the offspring parameters \(\varvec{\theta }_{\varphi }\) can be recovered similar to the results in the main text. Here, we only report the average test-likelihood \(\ell _\mathrm{test}\) of 12 unseen data sets and \(\ell _2\) norm between true background intensity and predicted \(\hat{\mu }\), see Table 9. GP-ETAS performs better in Experiment (1a) and (1b) compared with standard ETAS models.
Number of events assigned to the background process \(\mu (\varvec{x})\) in each iteration of the GP-ETAS Gibbs sampler. First to third column corresponds to Case 1, Case 2 and Case 3, as described in Sect. 5 of the main text. First row Experiment (1a), only one event is assigned as background event at the first iteration. Second row Experiment (1b), almost all events are assigned to the background process at the first iteration. After several iterations (\(<2000\)), the Gibbs sampler converges to the true number of background events given by a horizontal, solid, magenta line
Results of experiment 2. Inference results of Experiment (2) are summarized in Table 10. Here, synthetic data is simulated on a scaled spatial domain \(\tilde{\mathcal {X}}=[0,500]\times [0,500]\). The background intensity model \(\tilde{\mu }_1(\varvec{x})\) underlying the simulated data can be retrieved in an acceptable way for all different initial values of the length scales \(\nu _1\), \(\nu _2\). Offspring parameters \(\varvec{\theta }_{\varphi }\) can also be retrieved in an acceptable way. We only report following metrics \(\ell _2\) (error in estimation the background intensity) and \(\ell _\mathrm{test}\) (test-likelihood of unseen data), which are described in the main text (Sect. 5), see Table 10. GP-ETAS has a lower error (\(\ell _2\)) of the estimated background intensity and a higher predictive power (\(\ell _\mathrm{test}\)) compared with standard ETAS models.
1.5 E Run times
Run times of GP-ETAS are given in Table 11. All computations have been done on a iMac Pro 2017 with 32 GB RAM and a processor with 2.3 GHz.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Molkenthin, C., Donner, C., Reich, S. et al. GP-ETAS: semiparametric Bayesian inference for the spatio-temporal epidemic type aftershock sequence model. Stat Comput 32, 29 (2022). https://doi.org/10.1007/s11222-022-10085-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11222-022-10085-3