Abstract
As the volume of scientific literature expands rapidly, accurately gauging and predicting the citation impact of academic papers has become increasingly imperative. Citation counts serve as a widely adopted metric for this purpose. While numerous researchers have explored techniques for projecting papers’ citation counts, a prevalent constraint lies in the utilization of a singular model across all papers within a dataset. This universal approach, suitable for small, homogeneous collections, proves less effective for large, heterogeneous collections spanning various research domains, thereby curtailing the practical utility of these methodologies. In this study, we propose a pioneering methodology that deploys multiple models tailored to distinct research domains and integrates early citation data. Our approach encompasses instance-based learning techniques to categorize papers into different research domains and distinct prediction models trained on early citation counts for papers within each domain. We assessed our methodology using two extensive datasets sourced from DBLP and arXiv. Our experimental findings affirm that the proposed classification methodology is both precise and efficient in classifying papers into research domains. Furthermore, the proposed prediction methodology, harnessing multiple domain-specific models and early citations, surpasses four state-of-the-art baseline methods in most instances, substantially enhancing the accuracy of citation impact predictions for diverse collections of academic papers.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The rapid advancement of science and technology has led to a staggering increase in the number of academic publications produced globally each year (Zhu & Ban, 2018). In this ever-growing landscape, effectively evaluating the impact of research papers has become a critical issue (Castillo et al., 2007; Chakraborty et al., 2014; Li et al., 2019; Yan et al., 2011). Citation count, which measures the frequency with which a paper is referenced by other works, is widely recognized as the most prevalent metric for assessing the influence of academic papers, authors, and institutions (Bu et al., 2021; Cao et al., 2016; Lu et al., 2017; Redner, 1998; Stegehuis et al., 2015; Wang et al., 2021). Building upon the foundation of citation counts, numerous additional measures have been proposed to quantify research impact from various perspectives (Braun et al., 2006; Egghe, 2006; Garfield, 1972, 2006; Hirsch, 2005; Persht, 2009; Yan & Ding, 2010).
Predicting the impact of scientific papers has garnered significant research attention due to its profound implications (Abramo et al., 2019; Abrishami & Aliakbary, 2019; Bai et al., 2019; Cao et al., 2016; Chen & Zhang, 2015; Li et al., 2019; Liu et al., 2020; Ma et al., 2021; Ruan et al., 2020; Su, 2020; Wang et al., 2013, 2021, 2023; Wen et al., 2020; Xu et al., 2019; Yan et al., 2011; Yu et al., 2014; Zhao & Feng, 2022; Zhu & Ban, 2018). See “Citation count prediction” section for more detailed discussion about them. Accurately forecasting the future citation impact of academic papers, particularly those recently published, offers invaluable benefits to various stakeholders within the research ecosystem. Precisely predicting the impact of papers, especially those published for a short time, would be helpful for researchers to find potentially high-impact papers and interesting research topics at an earlier stage. It is also helpful for institutions, government agencies, and funding bodies to evaluate published papers, researchers, and project proposals, among others.
For large and diverse collections encompassing papers from various research areas, a one-size-fits-all approach to citation impact prediction may be inadequate. Even within a broad field like Computing, sub-fields such as Theoretical Computing, Artificial Intelligence, Systems, and Applications can exhibit distinct citation patterns. Previous study has demonstrated that citation dynamics can vary significantly across research areas, journals, researchers in different age groups, among other factors (Kelly, 2015; Levitt & Thelwall, 2008; Mendoza, 2021; Milz & Seifert, 2018). To illustrate this point, let us consider an example from the DBLP dataset used in our study. Figure 1a depicts the average citation distributions of papers in three research areas: Cryptography, Computer Networks, and Software Engineering. We can observe striking differences in their citation patterns:
-
Software Engineering papers consistently attract relatively few citations over time, without a pronounced peak in their citation curve.
-
Artificial Intelligence papers garner the highest citation counts among the three areas. Their citation curve rises rapidly, peaking around year 4, followed by a gradual decline until year 7, after which the decrease becomes more precipitous.
-
Cryptography papers exhibit a steadily increasing citation trend over the first 10 years, reaching a peak around year 11, followed by a slow decline in citations thereafter.
These divergent citation patterns across research areas highlight the limitations of employing a single, universal model for citation impact prediction. In light of these observations, a more effective strategy would be to segment papers into distinct groups based on their research areas and develop tailored prediction models for each group. By accounting for the unique citation characteristics of different domains, such a group-specific modelling approach has the potential to significantly enhance the accuracy and reliability of citation impact predictions, particularly for large and heterogeneous collections of academic papers.
Citation patterns are not solely determined by research areas but also influenced by the quality and intrinsic characteristics of individual papers. Even within the same research area, the citation dynamics of papers can vary considerably (Garfield, 2006; Wang et al., 2021; Yan & Ding, 2010). High-impact papers may exhibit significantly different citation trajectories compared to average or low-impact works. Accounting for these differences by employing multiple models tailored to papers with varying citation potential could further improve prediction performance. Figure 1b illustrates this phenomenon using an example from the Embedded & Real-Time Systems research area. All papers in this domain can be categorized into four classes based on their cumulative citation counts (cc) over 15 years: cc < 10, 10 ≤ cc < 50, 50 ≤ cc < 100, and cc ≥ 100. The general pattern observed for all the curves is that they initially increase for a few years and then decrease afterwards. However, the peak point varies depending on the total number of citations. Papers with higher citation counts take more years to reach their peak point. This finding suggests that class-based prediction can be a viable approach for our prediction task, as it account for the varying peak times based on the citation count classes.
If all of the papers are not classified, then it is necessary to have a classification system that encompasses multiple categories and an automated method for allocating each paper into one or more suitable categories. For a large collection of papers to be classified, both the effectiveness and efficiency of the allocating method are crucial factors to consider.
Taking into account all the observations mentioned earlier, we propose MM, a prediction method based on Multiple Models tailored for different research areas and citation counts, to predict the future citation counts of a paper. This work makes the following contributions:
-
1.
A new instance-based learning method is introduced to classify papers into a given number of research areas. Both paper contents (titles and abstracts) and citations are considered separately. An ensemble-based method is then employed to make the final decision. Experiments with the DBLP dataset demonstrate that the proposed method can achieve excellent classification performance.
-
2.
A prediction method for paper citation counts is proposed. For any paper to be predicted, a suitable prediction model is chosen based on its research area and early citation history. This customized approach enables each document to use a fitting model.
-
3.
Experiments with two datasets show that the proposed prediction method outperforms four baseline methods in this study, demonstrating its superiority.
The remainder of this article is structured as follows: “Related work” section reviews related work on citation count prediction and classification of academic papers. “Methodology” section describes the proposed method in detail. “Experimental settings and results” section presents the experimental settings, procedures, and results, along with an analysis of the findings. Finally, “Conclusion” section concludes the paper.
Related work
In this work, the primary task is citation count prediction of papers, while classification of scientific papers serves as an additional task that may be required for the prediction task. Accordingly, we review some related work on citation count prediction and classification of academic papers separately in the following sections.
Citation count prediction
In the literature, there are numerous papers on predicting the citation counts of scientific papers. These methods can be categorized into three groups based on the information used for prediction.
The first group relies solely on the paper’s citation history as input. Wang et al. (2013) developed a model called WSB to predict the total number of citations a paper will receive, assuming its earlier citation data is known. Cao et al. (2016) proposed a data analytic approach to predict the long-term citation count of a paper using its short-term (three years after publication) citation data. Given a large collection of papers C with long citation histories, for a paper p with a short citation history, they matched it with a group of papers in C with similar early citation data and then used those papers in C to predict p’s later citation counts. Abrishami and Aliakbary (2019) proposed a long-term citation prediction method called NNCP based on Recurrent Neural Network (RNN) and the sequence-to-sequence model. Their dataset comprised papers published in five authoritative journals: Nature, Science, NEJM (The New England Journal of Medicine), Cell, and PNAS (Proceedings of the National Academy of Sciences). Wang et al. (2021) introduced a nonlinear predictive combination model, NCFCM, that utilized multilayer perceptron (MLP) to combine WSB and an improved version of AVR for predicting citation counts.
The second group uses not only the citation data but also some other extracted features from the paper or the wider academic network for the prediction task. Yu et al. (2014) adopted a stepwise multiple regression model using four groups of 24 features, including paper, author, publication, and citation-related features. Bornmann et al. (2014) took the percentile approach of Hazen (1914), considering the journal’s impact and other variables such as the number of authors, cited references, and pages. Castillo et al. (2007) used information about past papers written by the same author(s). Chen and Zhang (2015) applied Gradient Boosting Regression Trees (GBRT) with six paper content features and 10 author features. Bai et al. (2019) made long-term predictions using the Gradient Boosting Decision Tree (GBDT) model with five features, including the citation count within 5 years after publication, authors’ impact factor, h-index, Q value, and the journal's impact factor. Akella et al. (2021) exploited 21 features derived from social media shares, mentions, and reads of scientific papers to predict future citations with various machine learning models, such as Random Forest, Decision Tree, Gradient Boosting, and others. Xu et al. (2019) extracted 22 features from heterogeneous academic networks and employed a Convolutional Neural Network (CNN) to capture the complex nonlinear relationship between early network features and the final cumulative citation count. Ruan et al. (2020) employed a four-layer BP neural network to predict the 5th year citation counts of papers, using a total of 30 features, including paper, author, publication, reference, and early citation-related features. By extracting high-level semantic features from metadata text, Ma et al. (2021) adopted a neural network to consider both semantic information and the early citation counts to predict long-term citation counts. Wang et al. (2023) applied neural network technology to a heterogeneous network including author and paper information. Huang et al. (2022) argued that citations should not be treated equally, as the citing text and the section in which the citation occurs significantly impact its importance. Thus, they applied deep learning models to perform fine-grained citation prediction—not just citation count for the whole paper but citation count occurring in each section.
The third group uses other types of information beyond those mentioned above. To investigate the impact of peer-reviewing data on prediction performance, Li et al. (2019) adopted a neural network prediction model, incorporating an abstract-review match method and a cross-review match mechanism to learn deep features from peer-reviewing texts. Combining these learned features with breadth features (topic distribution, topic diversity, publication year, number of authors, and average author h-index), they employed a multilayer perceptron (MLP) to predict citation counts. Li et al. (2022) also utilized peer-reviewing text for prediction, using an aspect-aware capsule network. Zhao and Feng (2022) proposed an end-to-end deep learning framework called DeepCCP, which takes an early citation network as input and predicts the citation count using both GRU and CNN, instead of extracting features.
Citation counts of a paper can be affected by many factors such as research areas, paper types, age, sex, and other aspects of the authors (Andersen & Nielsen, 2018; Mendoza, 2021; Thelwall, 2020). Levitt and Thelwall (2008) compared patterns of annual citations of highly cited papers across six research areas. To our knowledge, Abramo et al. (2019) is the only work that uses multiple regression models for prediction, with one model for each subject category. Abramo et al. (2019) is the most relevant to our work in this article. However, there are two major differences. First, we propose a paper classification method in this paper, while no paper classification is required in Abramo et al. (2019). Second, we apply multiple models for papers in each category, whereas only one model is used for each category in Abramo et al. (2019).
Classification of scientific papers
Classification of scientific papers becomes a critical issue when organizing and managing an increasing number of publications through computerized solutions. In previous research, typically, meta-data such as title, abstract, keywords, and citations of papers were used for this task, while full text was not considered due to its unavailability in most situations.
Various machine learning methods, such as K-Nearest Neighbors (Lukasik et al., 2013; Waltman & Van Eck, 2012), K-means (Kim & Gil, 2019), and Naïve Bayes (Eykens et al., 2021), have been applied. Recently, deep neural network models, such as Convolutional Neural Networks (Daradkeh et al., 2022; Rivest et al., 2021), Recurrent Neural Networks (Hoppe et al., 2021; Semberecki & Maciejewski, 2017), and pre-trained language models (Hande et al., 2021; Kandimalla et al., 2020), have also been utilized.
One key issue is the classification system to be used. There are many different classification systems. Both Thomson Reuters’ Web of Science database (WoS) and Elsevier’s Scopus database have their own general classification systems, covering many subjects/research areas. Some systems focus on one particular subject, such as the medical subject headings (MeSH), the physics and astronomy classification scheme (PACS), the Chemical Abstracts Sections, the journal of economic literature (JEL), and the ACM Computing Classification System.
Based on the WoS classification system, Kandimalla et al. (2020) applied a deep attentive neural network (DANN) to a collection of papers from the WoS database for the classification task. It was assumed that each paper belonged to only one category, and only abstracts were used.
Zhang et al. (2022) compared three classification systems: Thomson Reuters’ Web of Science, Fields of Research provided by Dimensions, and the Subjects Classification provided by Springer Nature. Among these, the second one was generated by machine learning methods automatically, while the other two were generated manually by human experts. It is found there are significant inconsistency between machine and human-generated systems.
Rather than using an existing classification system, some researchers build their own classification system using the collection to be classified or other resources such as Wikipedia.
Shen et al. (2018) organized scientific publications into a hierarchical concept structure of up to six levels. The first two levels (similar to areas and sub-areas) were manually selected, while the others were automatically generated. Wikipedia pages were used to represent the concepts. Each publication or concept was represented as an embedding vector, thus the similarity between a publication and a concept could be calculated by the cosine similarity of their vector representations. It is a core component for the construction of the Microsoft Academic Graph.
In the same vein as Shen et al. (2018), Toney-Wails and Dunham (2022) also used Wikipedia pages to represent concepts and build the classification system. Both publications and concepts were represented as embedding vectors. Their database contains more than 184 million documents in English and more than 44 million documents in Chinese.
Mendoza et al. (2022) presented a benchmark corpus and a classification system as well, which could be used for the academic paper classification task. The classification system used is the 36 subjects defined in the UK Research Excellent Framework.Footnote 1 According to Cressey and Gibney (2014), this practice is the largest overall assessment of university research outputs ever undertaken globally. The 191,000 submissions to REF 2014 comprise a very good data set because every paper was manually categorized by experts when submitted.
Liu et al. (2022) described the NLPCC 2022 Task 5 Track 1, a multi-label classification task for scientific literature, where one paper may belong to multiple categories simultaneously. The data set, crawled from the American Chemistry Society’s publication website, comprises 95,000 papers’ meta-data including titles and abstracts. A hierarchical classification system, with a maximum of three levels, was also defined.
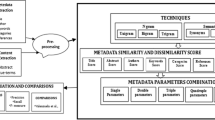
As we can see, the classification problem of academic papers is quite complicated. Many classification systems and classification methods are available. However, classification systems and classification methods are related to each other. The major goal of this work is to perform citation count prediction of published papers, in which classification of papers is a basic requirement. For example, considering the DBLP dataset which includes over four million papers, special consideration is required to perform the classification task effectively and efficiently. We used the classification system from CSRankings,Footnote 2 which included a set of four categories (research areas) and 26 sub-categories in total. A group of top venues were identified for each sub-category. However, many more venues in DBLP are not assigned to any category. We used all those recommended venue papers in the CSRankings system as representative papers of a given research area. An instance-based learning approach was used to measure the semantic similarity of the target paper and all the papers in a particular area. A decision could be made based on the similarity scores that the target paper obtained for all research areas. Besides, citation data between the target paper and all the papers in those recommended venues is also considered. Quite different from those proposed classification methods before, this instance-based learning approach suits our purpose well. See “Methodology” section for more details.
Methodology
This research aims to predict the number of citations of academic papers in the next couple of years based on their metadata including title, abstract and citation data since publication. The main idea of our approach is: for a paper, depends on its research area and early citation count, we use a specific model to make the prediction. There are two key issues. Academic paper classification and citation count prediction methods. Let us detail them one by one in the following subsections.
Computing classification system
To carry out the classification task of academic papers, a suitable classification system is required. There are many classification systems available for natural science, social science, humanities, or specific branches of science or technology. Since one of the datasets used in this study is DBLP, which includes over four million papers on computer science so far, we will focus our discussion on classification systems and methods for computer science.
In computer science, there are quite a few classification systems available. For example, both the Association for Computing Machinery (ACM) and the China Computer Federation (CCF) define their own classification systems. However, both are not very suitable for our purpose. The ACM’s classification system is quite complicated, but it does not provide any representative venues for any of the research areas. The CCF defines 10 categories and recommends dozens of venues in each category. However, some journals and conferences publish papers in more than one category, but they are only recommended in one category. For instance, both the journals IEEE Transactions on Knowledge and Data Engineering and Data and Knowledge Engineering publish papers on Information Systems and Artificial Intelligence, but they are only recommended in the Database/Data Mining/Content Retrieval category.
In this research, we used the classification system from CSRankings. This system divides computer science into four areas: AI, System, Theory, and Interdisciplinary Areas. Then, each area is further divided into several sub-areas, totalling 26 sub-areas. We flatten these 26 sub-areas for classification, while ignoring the four general areas at level one. One benefit of using this system is that it lists several key venues for every sub-area. For example, three venues are identified for Computer Vision: CVPR (IEEE Conference on Computer Vision and Pattern Recognition), ECCV (European Conference on Computer Vision), and ICCV (IEEE International Conference on Computer Vision). This is very useful for the paper classification task, as we will discuss now.
Paper classification
For this research, we need a classification algorithm that can perform the classification task for all the papers in the DBLP dataset effectively and efficiently.
Although many classification methods have been proposed, we could not find a method that suits our case well. Therefore, we developed our own approach. Using the classification system of CSRankings, we assume that all the papers published in those identified venues belong to that given research area, referred to as seed papers. For all the non-seed papers, we need to decide the areas to which they belong. This is done by considering three aspects together: content, references, and citations. Let us look at the first aspect first.
The collection of all the seed papers, denoted as C, was indexed using the Search engine LuceneFootnote 3 with the BM25 model. Both titles and abstracts were used in the indexing process. Each research area \({a}_{k}\) is presented by all its seed papers C(\({a}_{k}\)). For a given non-seed paper p, we use its title and abstract as a query to search for similar papers in C. Then each seed paper s will obtain a score (similarity between p and s)
in which b1 and b2 are two parameters (set to 0.75 and 1.2, respectively, as default setting values of Lucene in the experiments), Ts is the set of all the terms in s, \(AL(C)\) is the average length of all the documents in C, \(f\left({t}_{j},s\right)\) is the term frequency of \({t}_{j}\) in s, \(idf\left({t}_{j}\right)\) is the inverse document frequency of \({t}_{j}\) in collection C with all the seed papers. \(idf\left({t}_{j}\right)\) is defined as
in which \(\left|C\right|\) is the number of papers in \(C\), and \(\left|C({t}_{j})\right|\) is the number of papers in C satisfying the condition that \({t}_{j}\) appears in them. For a paper p and a research area \({a}_{k}\), we can calculate the average similarity score between p and all the seed papers in C(\({a}_{k}\)) as
where C(\({a}_{k}\)) is the collection of seed papers in area \({a}_{k}\).
We also consider citations between \(p\) and any of the papers in C. Citations in two different directions are considered separately: \(citingNum\left( p,{a}_{k}\right)\) denotes the number of papers in C(\({a}_{k}\)) that p cites, and \(citedNum\left(p,{a}_{k}\right)\) denotes the number of papers in C(\({a}_{k}\)) that cites p. Now we want to combine the three features. Normalization is required. For example, \(sim\left({p,a}_{k}\right)\) can be normalized by
in which \(RA\) is the set of 26 research areas. \(citingNum\left(p,{a}_{k}\right)\) and \(citedNum\left(p,{a}_{k}\right)\) can be normalized similarly. Then we let
for any \({a}_{k}\in RArea\), in which \({\beta }_{1}\), \({\beta }_{2}\), and \({\beta }_{3}\) are three parameters. When applying Eq. 5 to \(p\) and all 26 research areas, we may obtain corresponding scores for each area. p can be put to research area \({a}_{k}\) if \(score\left(p,{a}_{k}\right)\) is the biggest among all 26 scores for all research areas. The values of \({\beta }_{1}\), \({\beta }_{2}\), and \({\beta }_{3}\) are decided by Euclidean Distance with multiple linear regression with a training data set (Wu et al., 2023). Compared with other similar methods such as Stacking with MLS and StackingC, this method can achieve comparable performance but much more efficient than the others. It should be very suitable for large-scale datasets.
In this study, we assume that each paper just belongs to one of the research areas. If required, this method can be modified to support multi-label classification, then a paper may belong to more than one research area at the same time. We may set a reasonable threshold \(\tau\), and for any testing paper \(p\) and research area \({a}_{k}\), if \(score\left(p,{a}_{k}\right)>\tau\), then paper \(p\) belongs to research area \({a}_{k}\). However, this is beyond the scope of this research, and we leave it for further study.
In summary, the proposed classification algorithm instance-based learning (IBL) is sketched as follows:
Citation count prediction
As we observed that papers in the same research area may have different citation patterns, it is better to treat them using multiple prediction models rather than one unified model. Therefore, for all the papers in a research area, we divide them into up to 10 groups according to the number of citations already obtained in the first m years. In a specific research area, for a group of papers considered, we count the number of citations they obtained during a certain period. We use cc (i) to represent the number of papers cited i times, where i ranges from 0 to n.
A threshold of 100 is set. We consider the values of cc(0), cc(1),…, cc(n) in order. If cc(0) is greater than or equal to the threshold, we create a group with those papers that received zero citations. Otherwise, we combine cc(0) with cc(1), and if the sum is still less than the threshold, we continue adding the next value cc(2), and so on, until the cumulative sum reaches or exceeds the threshold. At this point, we create a group with all the papers contributing to that cumulative sum. We then move on to the next unassigned value of cc(i) and repeat the process, creating new groups until all papers are assigned to a group. The last group may contain fewer than 100 papers, but it is still considered a valid group.
A regression model is set for each of these groups for prediction. For the training data set, all the papers are classified by research area with known citation history of up to t years. For all the papers belonging to a group \({g}_{i}\) inside a research area \({a}_{k}\), we put their information together. Consider
\({c}_{0}\), \({c}_{1}\), …,\({c}_{m}\), and \({\text{c}}_{t}\) are citation counts of all the papers involved up to year 0, 1,…, m, and in year t (t ≥ m). We can train the weights \({\text{w}}_{0}\), \({\text{w}}_{1}\),…, \({\text{w}}_{m}\), and b for this group by multiple linear regression using \({c}_{0}\), \({c}_{1}\), …,\({c}_{m}\) as independent variables and \(c^{\prime}_{t}\) as the target variable. The same applies to all other groups and research areas.
To predict the future citation counts of a paper, we need to decide which research area and group that paper should be in. Then the corresponding model can be chosen for the prediction. Algorithm MM is sketched as follows:
Note that classification and citation count prediction are two separate tasks. When performing the citation count prediction task, it is required that all the papers involved should have a research area label. Such a requirement can be satisfied in different ways. For example, in the WoS system, it has a list of journals, and each journal is assigned to one or two research areas. All the papers published in those journals are classified by the journals publishing them. In arXiv, an open-access repository of scientific papers, all the papers are assigned a research area label by the authors when uploading them. When performing the citation count prediction task on such datasets, we do not need to do anything else. However, for papers in DBLP, all the papers are not classified. It is necessary to classify them in some way before we can perform citation count prediction for all the papers involved. In this study, we proposed an instance-based learning approach, which provides an efficient and effective solution to this problem.
Experimental settings and results
Datasets
Two datasets were used for this study. One is a DBLP dataset, and other is an arXiv dataset.
We downloaded a DBLP dataset (Tang et al., 2008).Footnote 4 It contains 4,107,340 papers in computer science and 36,624,464 citations from 1961 to 2019. For every paper, the dataset provides its metadata, such as title, abstract, references, authors and their affiliations, publication year, the venue in which the paper was published, and citations since publication. Some subsets of it were used in this study.
For the classification part, we used two subsets of the dataset. The first one (C1) is all the papers published in those 72 recommended venues in CSRankings between 1965 and 2019. There are 191,727 papers. C1 is used as seed papers for all 26 research areas. The second subset (C2) includes 1300 papers, 50 for each research area. Those papers were randomly selected from a group of 54 conferences and journals and judged manually. C2 is used for the testing of the proposed classification method.
For the prediction part, we also used two subsets. One subset for training and the other for testing. The training dataset (C3) includes selected papers published between 1990 and 1994, and the testing dataset (C4) includes selected papers published in 1995. For all those papers between 1990 and 1994 or in 1995, we removed those that did not get any citation and those with incomplete information. After such processing, we obtain 38,247 papers for dataset C3, and 9967 papers for dataset C4.
We also downloaded an arXiv dataset (Saier and Farber, 2020).Footnote 5 It contains 1,043,126 papers in many research areas including Physics, Mathematics, Computer Science, and others, with 15,954,664 citations from 1991 to 2022. For every paper, its metadata such as title, abstract, references, authors and affiliations, publication year, and citations since publication was provided. Importantly, each paper is given a research area label by the authors. Therefore, it is no need to classify papers when we use this dataset for citation count prediction. Two subsets were generated in this study. One subset for training and the other for testing. The training dataset (C5) includes all the papers published between 2008 and 2013, and the testing dataset (C6) includes all the papers published in 2014. There are 5876 papers in dataset C5 and 1471papers in dataset C6.
Classification results
In the CSRankings classification system, there are a total of 26 special research areas. A few top venues are recommended for each of them. We assume that all the papers published in those recommended conferences belong to the corresponding research area solely. For example, three conferences CVPR, ECCV, and ICCV are recommended for Computer Vision. We assume that all the papers published in these three conferences belong to the Computer Vision research area but no others.
To evaluate the proposed method, we used a set of 1300 non-seed papers (C2). It included 50 papers for each research area. All of them were labelled manually. In Eq. 5, three parameters need to be trained. Therefore, we divided those 1300 papers into two equal partitions of 650, and each included the same number of papers in every research area. Then the two-fold cross-validation was performed. Table 1 shows the average performance.
We can see that the proposed method with all three features, content similarity (Sim), citation to other papers (To evaluate the proposed method, we used a set of 1300 non-seed papers (C2). It included 50 papers for each research area. All of them were labelled manually. In Eq. 5, three parameters need to be trained. Therefore, we divided those 1300 papers into two equal partitions of 650, and each included the same number of papers in every research area. Then the two-fold cross-validation was performed. Table 1 shows the average performance.
We can see that the proposed method with all three features, content similarity (\(sim\)), citation to other papers (\(citingNum\)), and citation by others (\(citedNum\)), are useful for the classification task. Roughly citation in both directions (\(citingNum+citedNum\)) and content similarity (\(sim\)) have the same ability. Considering three features together, we can obtain an accuracy, or an F-measure, of approaching 0.8. We are satisfied with this solution. On the one hand, its classification performance is good compared with other methods in the same category, e.g., (Ambalavanan & Devarakonda, 2020; Kandimalla et al., 2020). In Kandimalla et al. (2020), F-scores across 81 subject categories are between 0.5 and 0.8 (See Fig. 1 in that paper). In Ambalavanan and Devarakonda (2020), the four models ITL, Cascade Learner, Ensemble-Boolean, and Ensemble-FFN obtain an F-score of 0.553, 0.753, 0.628, and 0.477, respectively, on the Marshall dataset they experimented with (see Table 4 in their paper). Although those results may not be comparable since the datasets used are different, it is an indicator that our method is very good. Besides, our method can be implemented very efficiently. When the seed papers are indexed, we can deal with a large collection of papers very quickly with very little resource. The method is very scalable.
Setting for the prediction task
For the proposed method MM, we set 10 as the number of groups in each research area for the DBLP dataset, and 5 for the arXiv dataset. This is mainly because the arXiv dataset is smaller and has fewer papers in each research area.
Apart from MM, five baseline prediction methods were used for comparison:
-
1.
Mean of early years (MEY). It is a simple prediction function which returns the average of early citations of the paper as its predicted citations in the future (Abrishami & Aliakbary, 2019).
-
2.
AVR. Assume that there is a collection of papers with known citation history as the training data set. For a given paper for prediction, this method finds a group of most similar papers in the training set relating to their early citations (with the minimal sum of the squared citation count difference over the years), and then utilizes the average citations of those similar papers in the subsequent years as the predicted citation counts of the paper (Cao et al., 2016).
-
3.
RNN adopts a Recurrent Neural Network to predict papers’ future citation counts based on their early citation data (Abrishami & Aliakbary, 2019).
-
4.
OLS. Linear regression is used for the prediction model (Abramo et al., 2019). There are four variants. Both OLS_res and OLS_log only use early citations as independent variables in their prediction models, while OLS2_res and OLS2_log use early citations and impact factors of journals in their prediction models. OLS_res and OLS2_res apply a linear regularization to their early citations, while OLS_log and OLS2_log apply a logarithmic regularization to their early citations.
-
5.
NCFCM adopts a neural network to predict papers’ future citation counts based on early citation data and two simple prediction model data (Wang et al., 2021).
Evaluation metrics
Two popular metrics are used to evaluate the proposed method and compare it with the baselines: mean square error (MSE) and the coefficient of determination (R2). For a given set of actual values Y \(=\{{y}_{1},{y}_{2},\dots ,{y}_{n}\}\) and set of predicted values \(\widehat{Y\boldsymbol{ }}=\{{\widehat{y}}_{1},{\widehat{y}}_{2},\dots ,{\widehat{y}}_{n}\}\), MSE and R2 are defined as follows:
where \({\overline{y} }_{i}\) is the average of all n values in y. MSE measures the variation of the predicted values from the actual values, thus smaller values of MSE are desirable. R2 measures the corelation between the predicted values and actual values, and its value is between 0 and 1, where R2 = 0 means no correlation, R2 = 1 means a perfect positive correlation between the predicted values and the actual values, thus larger values of R2 are desirable.
Evaluation results
Evaluation has been carried out on two different aspects: overall performance for all the papers and for 100 highly citated papers.
Overall prediction performance
For papers with 0–5 years of citation history, we predict their citation counts in three continuous years in the future. The results are shown in Tables 2, 3, 4, 5, 6, 7. “Zero years of early citation data” means that the prediction was made in the same year as the paper was published. “One year of early citation data” means that the prediction was made in the next year as the paper was published. The number in bold indicates the best performance.
One can see that MM performs the best in most cases. In a few cases, OLS2_res performs the best. This is because OLS2_res considers both the paper’s early citation history and the journal’s impact factor, and the latter is not considered in any other method. In this way, it gives OLS2_res some advantages, especially when the citation history is very short. In a few cases, RNN performs the best on the arXiv dataset. In one case, AVE and NCFCM tied for first place in R2. Because linear regression is used in both OLS_res and MM, a comparison between them is able to show that dividing papers into multiple research areas is a very useful strategy for us to obtain better prediction performance. See “Ablation Study of MM” section for further experiments and analysis.
Prediction performance of highly cited papers
An important application of citation prediction is the early detection of highly cited papers (Abrishami & Aliakbary, 2019). Therefore, we evaluate the performance of the proposed method and its competitors in predicting highly cited papers. Based on the total citation counts in 2000 (DBLP) and in 2019 (arXiv), 100 most cited papers were selected for prediction. For all the papers involved, we compute the MSE values between the predicted citation counts and the actual citation counts of them. The results are shown in Table 8.
From Table 8, one can see that MM performs better than all the others, except when k = 0 (which means zero years of early citation data) OLS2_res performs slightly better than MM in the DBLP dataset. In all other cases, MM outperforms the competing methods.
Ablation study of MM
MM mainly incorporates two factors including research area and early citation counts into consideration. It is desirable to find how these two factors impact prediction performance. Another angle is the number of groups divided in each research area. To find out the impact of these features on prediction performance, we define some variants that implement none or one of the features of MM.
-
1.
MM-RA (RA). A variant of the MM algorithm that only considers research area but not early citation counts.
-
2.
MM-CC (CC). A variant of the MM algorithm that only considers early citation counts.
-
3.
MM-5. A variant of the MM algorithm that divides all the papers in the area into 5 instead of 10 groups.
-
4.
MEY. It is a simplest variant of MM. It considers neither research area nor early citation counts.
Now let us have a look at how these variants perform compared with the original algorithm. See Tables 9, 10, 11, 12, 13, 14, 15 for the results. It is not surprising that MM performs better than three variants of MM including RA, CC and MEY, while MEY, the variant with none of the two components, performs the worst in predicting the citation counts of papers. Such a phenomenon demonstrates that both components including research area and early citation counts are useful for prediction performance, either used separately or in combination. However, the usefulness of these two components is not the same. The performance of CC is not as good as RA when k = 0 and k = 1, but better than RA when k > 1. Understandably, this indicates that RA is a more useful resource than citation history when the citation history is short, but the latter becomes more and more useful when the citation history becomes longer.
When applying the standard MM algorithm, we divide all the papers in one research area into 10 groups based on the number of citations they obtain in the early years. MM-5 reduces the number of groups from 10 to 5 simply by combining two neighbouring groups to one. MM is better than MM-5 in most cases and on average. The difference between them is small in most cases. However, it is noticeable that MM-5 performs better than MM in two cases, mainly because the size of some of the groups is very small, and the prediction based on such small groups is not very accurate.
Impact of classification on MM
For the DBLP dataset, some papers were classified automatically through the venues in which they were published, while many others were classified through the classification method IBL. It would be interesting to make a comparison of these two groups when performing the prediction task. The results are shown in Table 16 We can see that the group of non-seed papers gets better perdition results than the group of seed papers by a clear margin in all the cases. This demonstrates that the two methods IBL and MM can work together well for achieving good prediction results. On the other hand, such a result is a little surprising. Why can the non-seed group perform better than the seed group? One major reason is for the citation count prediction task, MSE values and citation counts have a strong positive correlation. In this case, there are 2346 seed papers (C7), whose average citation count is 6.339, while there are 7621 non-seed papers (C8), whose average citation count is 3.085. These two groups are not directly comparable because of the difference in average citation counts. Note that C4 = C7 + C8 (see “Datasets” section for C4’s definition).
To make the comparison fair to both parties, we select a subgroup from each of them by adding a restriction: those papers obtain a citation count in the range of [10,20] by the year 2000. We obtain 318 papers (C9) from the seed paper group and 418 papers (C10) from the non-seed paper group. Coincidentally, the average citation counts for both sub-groups are the same: 13.443. This time, the two groups are in a perfect condition for a comparison. Table 17 shows the results. Not surprisingly, those MSE value pairs are very close. It demonstrates that for papers either classified by our classification algorithm IBL, or categorized by recommended top venues, the prediction is equally good. It also implies that IBL can perform the classification work properly.
Conclusion
Different from previous studies, this paper applies multiple models to predict the citation counts of papers in the next couple of years, and each model fits a special research area and early citation history of the paper in question. The rationale behind this is: in general, papers in different research areas and with different early citation counts have their own citation patterns. To verify the prediction performance of the proposed method, we have tested it with two datasets taken from DBLP and arXiv. The experimental results show that the proposed MM method outperforms all the baseline methods involved in most cases in two tasks: the overall prediction performance of a large collection of paper and prediction performance of a group of highly cited papers.
As an important component of prediction for research papers, we have also presented a novel instance-based learning model for classification of research papers. By predefining a small group of papers in each category, the proposed method can classify new papers very efficiently with good accuracy.
As our future work, we would incorporating other types of information such as publication venues and author information. Then, prediction performance can be further improved. Secondly, we plan to explore using some deep learning methods for research paper classification. For example, such methods can be used to compare the content similarity of two research papers.
References
Abramo, G., D’Angelo, C., & Felici, G. (2019). Predicting publication long-term impact through a combination of early citations and journal impact factor. Journal of Informetrics, 13(1), 32–49.
Abrishami, A., & Aliakbary, S. (2019). Predicting citation counts based on deep neural network learning techniques. Journal of Informetrics, 13(2), 485–499.
Akella, A., Alhoori, H., Kondamudi, P., et al. (2021). Early indicators of scientific impact: Predicting citations with altmetrics. Journal of Informetrics, 15(2), 101128.
Ambalavanan, A. K., & Devarakonda, M. V. (2020). Using the contextual language model BERT for multi-criteria classification of scientific articles. Journal of Biomedical Informatics, 112, 103578.
Andersen, J. P., & Nielsen, M. W. (2018). Google Scholar and Web of Science: Examining gender differences in citation coverage across five scientific disciplines. Journal of Informetrics, 12(3), 950–959.
Bai, X., Zhang, F., & Lee, I. (2019). Predicting the citations of scholarly paper. Journal of Informetrics, 13(1), 407–418.
Bornmann, L., Leydesdorff, L., & Wang, J. (2014). How to improve the prediction based on citation impact percentiles for years shortly after the publication data? Journal of Informetrics, 8(1), 175–180.
Braun, T., Glänzel, W., & Schubeert, A. (2006). Hirsch-type index for journals. Scientometrics, 69(1), 169–173.
Bu, Y., Lu, W., Wu, Y., Chen, H., & Huang, Y. (2021). How wide is the citation impact of scientific publications? A cross-discipline and large-scale analysis. Information Processing & Management, 58(1), 102429.
Cao, X., Chen, Y., & Liu, K. (2016). A data analytic approach to quantifying scientific impact. Journal of Informetrics, 10(2), 471–484.
Castillo, C., Donato, D., & Gionis, A. (2007). Estimating number of citations using author reputation. String processing and information retrieval (pp. 107–117). Berlin: Springer.
Chakraborty, T., Kumar, S., Goyal, P., Ganguly, N., & Mukherjee, A. (2014). Towards a stratified learning approach to predict future citation counts. In IEEE/ACM joint conference on digital libraries (pp. 351–360). IEEE.
Chen, J., & Zhang, C. (2015). Predicting citation counts of papers. In 2015 IEEE 14th international conference on cognitive informatics & cognitive computing (ICCI* CC) (pp. 434–440). IEEE.
Cressey, D., & Gibney, E. (2014). UK releases world’s largest university assessment. Nature. https://doi.org/10.1038/nature.2014.16587
Daradkeh, M., Abualigah, L., Atalla, S., & Mansoor, W. (2022). Scientometric analysis and classification of research using convolutional neural networks: A case study in data science and analytics. Electronics, 11(13), 2066.
Egghe, L. (2006). Theory and practice of the g-index. Scientometrics, 69(1), 131–152.
Eykens, J., Guns, R., & Engels, T. (2021). Fine-grained classification of social science journal articles using textual data: A comparison of supervised machine learning approaches. Quantitative Science Studies, 2(1), 89–110.
Garfield, E. (1972). Citation analysis as a tool in journal evaluation. Science, 178(4060), 471–479.
Garfield, E. (2006). The history and meaning of the journal impact factor. JAMA, 295(1), 90–93.
Hande, A., Puranik, K., Priyadharshini, R., & Chakravarthi, B. (2021). Domain identification of scientific articles using transfer learning and ensembles. PAKDD, 2021, 88–97.
Hazen, A. (1914). Storage to be provided in impounding reservoirs for municipal water supply. Transactions of American Society of Civil Engineers, 77(1914), 1539–1640.
Hirsch, J. (2005). An index to quantify an individual’s scientific research output. Proceedings of the National Academy of Science of the United States of America, 102(46), 16569–16572.
Hoppe, F., Dessì, D., & Sack, H. (2021). Deep learning meets knowledge graphs for scholarly data classification. WWW (companion Volume), 2021, 417–421.
Huang, S., Huang, Y., Bu, Y., et al. (2022). Fine-gained citation count prediction via a transformer-based model with among-attention mechanism. Information Processing & Management, 59(2), 102799.
Kandimalla, B., Rohatgi, S., Wu, J., & Lee Giles, C. (2020). Large scale subject category classification of scholarly papers with deep attentive neural networks. Frontiers in Research Metrics and Analytics, 5, 600382.
Kelly, M. (2015). Citation patterns of engineering, statistics, and computer science researchers: An internal and external citation analysis across multiple engineering subfields. College and Research Libraries, 76(7), 859–882.
Kim, S., & Gil, J. (2019). Research paper classification systems based on TF-IDF and LDA schemes. Human-Centric Computing and Information Sciences, 9, 30.
Levitt, J. M., & Thelwall, M. (2008). Patterns of annual citation of highly cited articles and the prediction of their citation ranking: A comparison across subjects. Scientometrics, 77(1), 41–60.
Li, S., Zhao, W. X., Yin, E. J., & Wen, J. R. (2019). A neural citation count prediction model based on peer review text. In Proceedings of the 2019 conference on empirical methods in natural language processing and the 9th international joint conference on natural language processing (EMNLP-IJCNLP) (pp. 4914–4924).
Li, S., Li, Y., Zhao, W., et al. (2022). Interpretable aspect-aware capsule network for peer review based citation count prediction. ACM Transaction on Information System, 40(1), 1–29.
Liu, L., Yu, D., Wang, D., et al. (2020). Citation count prediction based on neural Hawkes model. IEICE Transactions on Information and Systems, 103(11), 2379–2388.
Liu, M., Zhang, H., Tian, Y., et al. (2022). Overview of NLPCC2022 shared task 5 track 1: Multi-label classification for scientific literature. NLPCC, 2(2022), 320–327.
Lu, C., Ding, Y., & Zhang, C. (2017). Understanding the impact change of a highly cited article: A content-based citation analysis. Scientometrics, 112(3), 927–945.
Lukasik, M., Kusmierczyk, T., Bolikowski, L., & Nguyen, H. (2013). Hierarchical, multi-label classification of scholarly publications: Modifications of ML-KNN algorithm. Intelligent Tools for Building a Scientific Information Platform, 2013, 343–363.
Ma, A., Liu, Y., Xu, X., et al. (2021). A deep learning based citation count prediction model with paper metadata semantic features. Scientometrics, 126(2), 6803–6823.
Mendoza, Ó. E., Kusa, W., El-Ebshihy, A., Wu, R., Pride, D., Knoth, P., Herrmannova, D., Piroi, F., Pasi, G. & Hanbury, A. (2022). Benchmark for research theme classification of scholarly documents. In Proceedings of the third workshop on scholarly document processing (pp. 253–262).
Mendoza, M. (2021). Differences in citation patterns across areas, article types and age groups of researchers. Publications, 9(4), 47.
Milz, T., & Seifert, C. (2018). Who cites what in computer science? Analysing citation patterns across conference rank and gender. TPDL, 2018, 321–325.
Persht, A. (2009). The most influential journals: Impact factor and Eigenfactor. Proceedings of the National Academy of Sciences, 106(17), 6883–6884.
Redner, S. (1998). How popular is your paper? An empirical study of the citation distribution. European Physical Journal B, 4(2), 131–134.
Rivest, M., Vignola-Gagné, E., & Archambault, É. (2021). Article-level classification of scientific publications: A comparison of deep learning, direct citation and bibliographic coupling. PLoS ONE, 16(5), e0251493.
Ruan, X., Zhu, Y., Li, J., et al. (2020). Predicting the citation counts of individual papers via a BP neural network. Journal of Informetrics, 4(3), 101039.
Saier, T., & Färber, M. (2020). UnarXive: A large scholarly data set with publications’ full-text, annotated in-text citations, and links to metadata. Scientometrics, 125, 3085–3108.
Semberecki, P., & Maciejewski, H. (2017). Deep learning methods for subject text classification of articles. FedCSIS, 2017, 357–360.
Shen, Z., Ma, H., & Wang, K. (2018). A web-scale system for scientific knowledge exploration. ACL, 4, 87–92.
Stegehuis, C., Litvak, N., & Waltman, L. (2015). Predicting the long-term citation impact of recent publications. Journal of Informetrics, 9(3), 642–657.
Su, Z. (2020). Prediction of future citation count with machine learning and neural network. In 2020 Asia-Pacific conference on image processing, electronics and computers (IPEC) (pp. 101–104). IEEE.
Tang, J., Zhang, J., Yao, L., Li, J., Zhang, L., & Su, Z. (2008). Arnetminer: Extraction and mining of academic social networks. In Proceedings of the 14th ACM SIGKDD international conference on Knowledge discovery and data mining (pp. 990–998).
Thelwall, M. (2020). Gender differences in citation impact for 27 fields and six English-speaking countries 1996–2014. Quantitative Science Studies, 1(2), 599–617.
Toney, A., & Dunham, J. (2022). Multi-label classification of scientific research documents across domains and languages. In Proceedings of the third workshop on scholarly document processing (pp. 105–114).
Waltman, L., & van Eck, N. (2012). A new methodology for constructing a publication-level classification system of science. Journal of the American Society for Information Science and Technology, 63(12), 2378–2392.
Wang, B., Wu, F., & Shi, L. (2023). AGSTA-NET: Adaptive graph spatiotemporal attention network for citation count prediction. Scientometrics, 128(1), 511–541.
Wang, D., Song, C., & Barabasi, A. (2013). Quantifying long-term scientific impact. Science, 342(6154), 127–132.
Wang, K., Shi, W., Bai, J., et al. (2021). Prediction and application of article potential citations based on nonlinear citation-forecasting combined model. Scientometrics, 126(8), 6533–6550.
Wen, J., Wu, L., & Chai, J. (2020). Paper citation count prediction based on recurrent neural network with gated recurrent unit. In 2020 IEEE 10th international conference on electronics information and emergency communication (ICEIEC) (pp. 303–306). IEEE.
Wu, S., Li, J., & Ding, W. (2023). A geometric framework for multiclass ensemble classifiers. Machine Learning, 112(12), 4929–4958.
Xu, J., Li, M., Jiang, J., et al. (2019). Early prediction of scientific impact based on multi-bibliographic features and convolutional neural network. IEEE ACCESS, 7, 92248–92258.
Yan, R., Tang, J., Liu, X., Shan, D., & Li, X. (2011). Citation count prediction: learning to estimate future citations for literature. In Proceedings of the 20th ACM international conference on Information and knowledge management (pp. 1247–1252).
Yan, E., & Ding, Y. (2010). Weighted citation: An indicator of an article’s prestige. Journal of the American Society for Information Science and Technology, 61(8), 1635–1643.
Yu, T., Yu, G., Li, P. Y., & Wang, L. (2014). Citation impact prediction for scientific papers using stepwise regression analysis. Scientometrics, 101, 1233–1252.
Zhang, L., Sun, B., Shu, F., & Huang, Y. (2022). Comparing paper level classifications across different methods and systems: an investigation of Nature publications. Scientometrics, 127(12), 7633–7651.
Zhao, Q., & Feng, X. (2022). Utilizing citation network structure to predict paper citation counts: A deep learning approach. Journal of Informetrics, 16(1), 101235.
Zhu, X. P., & Ban, Z. (2018). Citation count prediction based on academic network features. In 2018 IEEE 32nd international conference on advanced information networking and applications (AINA) (pp. 534-541). IEEE.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Data collection, programming and analysis were performed by Fang Zhang. The first draft of the manuscript was written by Shengli Wu, and all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have no relevant financial and non-financial interests to disclose.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhang, F., Wu, S. Predicting citation impact of academic papers across research areas using multiple models and early citations. Scientometrics (2024). https://doi.org/10.1007/s11192-024-05086-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11192-024-05086-0