Abstract
Health is a representative domain data-driven research since health research data are growingly generated at a massive scale. There is an intuitive logic that the degree to which disease burden and the number of data resources align. In order to figure out disease-specific data sharing and reuse level, we took the number of data records and their citations in the scientific literature in the Data Citation Index platform as approximate indicators. The results indicated that only a small percentage (7.5%) of health data records had received documented citations by scientific publications. We find the level of data sharing and reuse varies across diseases. Our study suggested that the more socioeconomic burden and the more research funding, the more likely scientific data for diseases will be produced and made available. But such a correlation could not be observed for the activity of data reuse. Secondary reuse of scientific data is a complex behavior.
Similar content being viewed by others
Background
Due in large part to government mandates to make research data freely accessible, there are increasing numbers of data repositories being created worldwide and filled with data by researchers. With the ever-increasing amount of scientific data being produced and made available, the scientific community urgently need to know the clear picture of data sharing and reuse in order to measure the impact of research data on scientific research. Compared with scientific publications, there are relatively fewer quantitative studies on scientific data in the community of Scientometrics and Informetrics, mainly due to the lack of large-scale and robust databases for data records. Launched in 2012, the Data Citation Index (DCI) is now included within the Web of Science, allowing for the discovery of the shared research data and measurement of data reuse since the research data has been linked to the published literature.
Data citation by scientific publication is not only an approach to indicating the secondary use of scientific data, but also is very necessary in order to protect the rights and interests of data providers and ensure the traceability of research data. It is a feasible step for promoting the repeatability and reproducibility of scientific research. The behavior of data citation consists of formal citation and informal citation. The practice of formal data citation includes citing a dataset or data record, or data article (a short, digestible description of research data that have been made publicly available) in the cited references section of a publication. However, the fact of the real world is “the informal data citation in the main text of articles is far more common than formal data citations in the references of articles” (Park et al., 2018). Informal data citation is difficult to track, but is an important way for researchers to share and reuse data, especially in the fields of life science and medical field (Park & Wolfram, 2017). The goal of the DCI is to harvest both formal and informal data citations from the literature. By connecting data to the research it informs, DCI provides a platform for linking data with the scientific literature which was underpinned, supported, and strengthened by these data (Force & Robinson, 2014). Therefore, by investigating the DCI, we can track how the data were used in the literature and thus evaluate the impact of research data on scientific research. Research data considered for inclusion include data studies, data sets, and software deposited in a recognized repository. As of March 24th, 2021, the DCI stored data from 431 repositories, including 10,763,120 data sets, 1,319,689 data studies and 190,346 software.
Health is a representative domain data-driven research since health research data are growingly generated at a massive scale. There are increasing investigations and applications of research data resources to advance the understanding of health and disease (Shilo et al., 2020). There is an intuitive logic that the degree to which disease burden and the number of data resources align. In other words, the more burden of disease, the larger number of data resources ready for research. Measures of disease burden such as incidence, mortality, years of life lost (YLLs) and disability-adjusted life years (DALYs) are fundamental indicators that represent public health conditions. DALYs is a mainly measure for burden of disease and it can be considered/calculated as sum of YLLs due to premature mortality and years lived with disability (YLDs) due to the occurrence of disease. (WHO, 2018). The disease burden has been correlated with scientific outputs such as biomedical publications (Evans ert al., 2014), clinical trials (Atal et al., 2018), medical patents (Huang et al., 2019) and scientific investment such as research grants (Jung et al., 2019; Zhang et al., 2020). To measure the misalignment of health needs and resource allocation for individual diseases, the disease burden is also compared with a group of research attention and research investment measures which include the research coverage in the scientific literature, the clinical trial coverage and the received funding (Yao et al., 2015). The research data (e.g., dataset), often represented as digital research objects, and understood as an important scientific output or resource, plays more underpinning roles for biomedical discovery and data-powered health.Footnote 1 Whether or not there is the association between disease burden and degree of data sharing/reuse remains uninvestigated. In addition, even when data are shared, the metadata, expertise, technologies and infrastructure necessary for reuse are lacking. Most published data sets are scattered into ‘supplemental files’ that are often impossible for machines or even humans to find. So, to make data reusable requires investment. Research institutions and funders are suggested to invest 5% of research funds in ensuring data are reusable (Mons, 2020). So, it is also worth exploring whether or not there is the association between funding count and data reuse.
In order to figure out disease-specific data sharing and reuse level, we took the number of data records and their citations in scientific literature in the DCI platform as approximate indicators. Focusing on health research data, we try to figure out (1) the distribution of data records across individual diseases, (2) the most highly cited data records, and (3) the relationship between the incidence or DALYs and the number of data records and citations for individual diseases, to compare global health needs and data sharing/reuse efforts, and (4) the relationship between data sharing/reuse and diseases-related funding count, to investigate if there is an association between funding count and the extent of data reuse.
Methods
Data sources
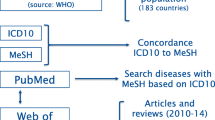
DCI is a database intend to facilitate the discovery of data, link data to paper, and encourage citation of data. DCI on the Web of Science was selected because DCI provides a single access to over 500 data repositories worldwide and to over two million data studies and data sets across multiple disciplines and monitors quality research data through a peer review process (Park & Wolfram, 2017). A study showed that “Thomson Reuters developed this database in response to a stated desire among members of the research community for increased attribution of non-traditional scholarly output such as data records.” Evaluation of data records can be realized through DCI, and our research can explore data records' relevance to scientific and scholarly research (Force & Robinson, 2014). While the reference of informal data records is not recorded in it, the platform can be used to explore the formal citations by scientific publications of data records (Park et al., 2018). As this study focuses on exploring the usage of clinical related data records, "Life Sciences & Biomedicine", one of the five research domains of Web of Science, is selected for searching DCI platform. Among them, 57 subject categories have been obtained in this domain on August 7th, 2020, and 39 discipline categories related to Life Sciences & Biomedicine were obtained after 18 discipline categories without data were removed. In total, 13,153 health-related data records were included in our study after removing the records related to molecular biology/pharmacy/animal. The workflow is shown in Fig. 1.
Data processing
We adopted the Medical Text Indexer (MTI) produced by the National Library of Medicine (NLM) to extract standardized medical subject headings terms from the title and abstract of data records in DCI. We focused on the diseases related MeSH terms.
According to the ICD-10 and MeSH terms mapping results, there are 115 diseases involving 6032 data records, accounting for 45.86% of all 13,153 health related data records extracted. Within the shared data records, there are 77 diseases matched with the corresponding data of funding amount, and among which 38 diseases related data records have been cited by scientific publications. The data processing process is shown in Fig. 1.
We used a correspondence table (Yegros-Yegros et al., 2020) to map World Health Organization (WHO) International Classification of Diseases (ICD-10) with MeSH terms. In order to evaluate the relationship between the data sharing, data reuse and health demand at the disease level, the number of data records related to diseases (indicating the level of data sharing), the citations by scientific publications (indicating the level of data reuse, that is, using them to generate new research) and the incidence of diseases (the social burden of diseases) were analyzed respectively. Incidence is defined as the number of new cases of a given cause during a given period in a specified population, which is an index representing the intensity of disease. We adopted DALYs, one of the Global Burden of Disease indicators provided by WHO, as the index for disease burden assessment. All the data were obtained from the Institute for Health Metrics and Evaluation (IHME2019) (Rubin, 2017).
Concerning the funding for diseases, we leveraged a data source in Global Observatory on Health R&D, which collected data on diseases/conditions in the number of grants for biomedical research by funder, type of grant, duration and recipients (WHO, 2019). Initiated by WHO in 2014 (Terry Robert et al., 2014). A global map of health R&D activity would improve the coordination of research and help to match limited resources with public health priorities. based on developments in semantic classification, and with better reporting of funded research though the Internet, it is now becoming feasible to create a global observatory for health R&D. This report provided statistics related to funding from 14 funding agencies that reported data to World RePORT (worldreport.nih.gov/). World RePORT is hosted by the U.S. National Institutes of Health and managed through a steering committee of the agencies providing data. To maintain World RePORT as open-access to the entire research community, especially those conducting research in low-and middle-income countries, this effort has been sponsored by the United States National Institutes of Health (NIH), the United Kingdom Medical Research Council (MRC), the Bill & Melinda Gates Foundation (BMGF), the European Commission (EC), the Canadian Institutes of Health Research, and the Wellcome Trust. World RePORT includes research projects supported by the following funding organizations. Bill & Melinda Gates Foundation (BMGF),Canadian Institutes of Health Research (CIHR), European Commission (EC), European & Developing Countries Clinical Trials Partnership (EDCTP), German Federal Ministry of Education and Research (BMBF), Global Alliance for Chronic Diseases (GACD), Japan Agency for Medical Research and Development (AMED), Medical Research Council (MRC), National Institutes of Health (NIH), Institut Pasteur, Swedish International Development Cooperation Agency (Sida), Swedish Research Council (SRC), United States Agency for International Development (USAID) and Wellcome Trust. We collected data on the classification of disease/condition in the report as funding counts.Footnote 2
Statistical analysis
SPSS 20.0 was used for statistical analysis of all the data. After single sample K-S test, the data of publications, citations and incidence/DALYs/fundings of various diseases were did not conform to the normal distribution, and Spearman Correlation Analysis was conducted to analyze the correlation among variables. Two-sided probability is taken as inspection level. P < 0.05 is statistically significant.
Results
Overall distribution of the data records
We retrieved a total of 13,153 clinical related data records, which first appeared in 1900. Data records are mainly concentrated in the recent 5 years. According to the classifications in DCI, we divided the data records into three types: 10,713 (81.45%) data sets, 1939 (14.74%) data studies and 60 (0.46%) repositories. Another 441(3.35%) records are software. Among all the data records included in our study, only 982 (7.5%) data records were cited by scientific publications. In other words, new studies were produced through the reuse of these data records (Fig. 2).
We could find the significant difference on the distribution of citations in clinical related data records according to Table 1. Although the repositories type has the lowest proportion of the number of data records, yet has the highest citation rate. Almost 85% of repositories have been cited by scientific publications, and this rate is higher than that of data studies (24.2%) and data sets (4%). And the number of average cited times for repositories is the highest. Data repository is defined as a database or collection comprising data studies, and data sets which stores and provides access to the raw data. Thus, repositories have more citable units to attract citations than individual data sets or software. To a certain extent, it shows that the sharing level of data sets and data studies is higher. Meanwhile, the reuse level of data repositories is higher, which has a greater impact on new research. Due in large part to government mandates to make research data freely accessible, there are increasing numbers of data repositories being created worldwide and filled with data by researchers.
Top 20 related diseases by the number of data records and their citations
There are some differences for the top 20 related diseases by the number of data records and their cited times by scientific publications respectively. Such diseases as chronic obstructive pulmonary disease, lower respiratory infections and Infectious bowel disease ranked in the top 10 output of data records, yet out of the top 20 list of reuse of data records. To some extent, the data records related to the above three diseases are broadly shared, but their reuse is relatively poor. The number of shared data records of bipolar disorder and inflammatory bowel disease, which list within the top 10 reuse, but is not in top 20 number of sharing. In addition, the rate of cited data records for such diseases as alcohol use disorders (14, 51.85%), acute hepatitis C (10, 30.30%), brain and nervous system cancers (8, 21.05%) and breast cancer (6, 25.00%), are over 20%, whereas they are not in the top 20 output diseases (Table 2).
The top 10 highly cited data records
In Table 3, we can see that 8 of the top 10 cited data records are listed as repositories, but these seem to be individual studies, not large-scale repositories, unless the data are longitudinal, and are not considered as individual data sets. Note that here we treat data sets, data studies, software and repositories (such as a Swiss HIV Cohort study) independently and thus compare them with each other. But for such repositories as Dryad, Fig share, they could not be treated as an independent data record since the data sets, data studies, software stored in these big platforms can be regarded as an independent data record.
Most of the top 10 cited data records are longitudinal cohort studies, which are often carried out for specific groups or regions. The research areas involved in these data records are mainly infectious diseases, aging study, family study and primary health care, from organizations and institutions in Europe and the United States. The most cited data record among them is Longitudinal Aging Study Amsterdam, which has been cited by 765 scientific publications (Table 3). We believe that strictly speaking the repositories listed in Table 3 are not representatives of related data records which are referenced together, but are large scale repositories that index potentially thousands of data records.
The relationship between data sharing/reuse and DALYs
According to our statistical results, all shared data records were related to 114 diseases and directly linked to 6032 data records, and the funding data for 77 diseases can be found. Among them, such data records which has been cited by scientific publications involved 51 diseases and included 369 data records, and the funding data for 38 diseases can be found. After single sample K-S test, the data of publications, citations and incidence/DALYs/funding count of various diseases did not conform to the normal distribution, so Spearman Correlation Analysis was conducted to analyze the correlation among variables. Table 4 showed the results of Spearman correlation analysis for each of two indicator pairs. It can be seen that there is no significant correlation between the sharing of disease-related data records and the disease incidence intensity. However, a moderate positive correlation can be observed for the efforts of data sharing with the DALYs and the funding of diseases.
The relationship between shared data records and funding count/DALYs for 77 diseases
The size of scatter points in Fig. 3 indicates the value of DALYs. The number of publications represents data sharing and citations corresponds to data reuse. According to the result of K-S normality test, the data with related relations were represented by scatter diagram in Figs. 3 and 4.
DM diabetes mellitus, T, B, LC Trachea, bronchus, lung cancers, COPD chronic obstructive pulmonary disease, SD skin diseases, LRI lower respiratory infections, RA rheumatoid arthritis, SCH schizophrenia, EP epilepsy, HHD hypertensive heart disease, CRC colon and rectum cancers, MOSC melanoma and other skin cancers, MS multiple sclerosis, ADD Alzheimer's disease and other dementias, Leukaemia leukaemia, Migraine migraine, KRU kidney, renal pelvis and ureter cancer, IHD ischaemic heart disease, BD bipolar disorder, AD anxiety disorders, CUC1 cervix uteri cancer, CME cardiomyopathy, myocarditis, endocarditis, PD1 Parkinson's disease, SC stomach cancer, BNSC brain and nervous system cancers, MD macular degeneration, LC liver cancer, OC2 ovary cancer, PC pancreas cancer, AUD alcohol use disorders, BC2 breast cancer, WC whooping cough, CUC2 corpus uteri cancer, OM otitis media, URE uncorrected refractive errors, BC1 bladder cancer, ED eating disorders, AAS autism and asperger syndrome, DD diarrhoeal diseases, CHA congenital heart anomalies, OC1 oesophagus cancer, PD2 periodontal diseases, CD chagas disease, IID idiopathic intellectual disability, PBC preterm birth complications, SCDT sickle cell disorders and trait, TC thyroid cancer, GBTC gallbladder and biliary tract cancer, URI upper respiratory infections, FBT food-bourne trematodes, YF yellow fever, DC dental caries, BNP back and neck pain, IDA iron-deficiency anaemia, PEM protein-energy malnutrition
In Fig. 3, We have found that the total number of disease-related data records were positively correlated with their funding overall. But we can found some diseases are in high incidence or funding count while have a low data record sharing level, such as the Malaria, Diarrhoeal diseases and protein-energy malnutrition. Some diseases have high DALYs and data sharing level while have a relative low level of funding support, such as Migraine.
The relationship between reused data records and funding count for 38 diseases
In Fig. 4, some diseases with a higher level of funding support and higher incidence intensity yet have a low level of data citation, such as Ischemic heart disease, and Lower respiratory infections. Some diseases with high incidence and relatively large number of citations have low level of funding support, such as migraine.
Discussions and conclusions
First, we give a basic analysis about the level of health data sharing and reuse through DCI. Our study revealed that data sets are the predominate type of disease-related data sharing on DCI platform, followed by data studies and repositories, while repositories contributed the most majority of data reuse. The overwhelming majority of data records are published in the recent five years and the proportion is 76.07%. Our study also gives a report on the impact of health-related data as measured by data citations in the DCI. The results indicated that only a small percentage (7.5%) of health data records had received documented citations by scientific publications. We find the level of data sharing and reuse varies across diseases, just like the imbalance of clinical trials across diseases found by (Marshall et al., 2021). Most recently, Zuo et al. (2021) provided a review on novel coronavirus disease (COVID-19) datasets extracted from PubMed Central articles. They found that 28.5% of 12,324 COVID-19 full-text articles in PubMed Central provided at least one dataset link. Epidemiological datasets accounted for the largest portion (53.9%) in the dataset collection, and most datasets (84.4%) were available for immediate download. PubMed Central articles are an important source of COVID-19 datasets, but there is significant heterogeneity in the way these datasets are mentioned, shared, updated and cited. Efforts to make scientific research results open and reproducible are increasingly reflected by journal policies, in which authors were encouraged or mandated to provide data availability statements. As a consequence of this policy, there has been a strong uptake of data availability statements in recent literature (Colavizza et al., 2020).
Previous studies have shown that the burden of disease is not always strongly correlated with the amount of funds and scientific publications related to disease, for example, some underfunded or overfunded diseases can be found. And underactive or overactive disease areas can be also found (Yegros-Yegros et al., 2020; Zhang et al., 2020). Work by Evans et al. found research attention and disease burden aligned within countries, but not at the global scale (Evans et al., 2014). Our work is built on the global data of disease burden and the number of data sharing and citations. Our study suggested that the more socioeconomic burden and the more research funding, the more likely scientific data for diseases will be produced and made available. But such a correlation could not be observed for the activity of data reuse. Secondary reuse of scientific data is a complex behavior. The University of Tennessee in the United States conducted an international survey in 2011 (Tenopir et al., 2011). From a disciplinary perspective, compared with environmental sciences, social sciences, physics, chemistry, and biology, medical researchers with the most unwilling reuse their own data, but with the most willing to use the data shared by others to conduct research or apply for projects. The main reasons why scientists are unwilling to share scientific data are time consuming, lack of funding and lack of standardization of data citations (worries about not being recognized after sharing). If certain conditions are met such as formal citation respondents agree they are willing to share their data. Our research indicated that the amount of health data sharing and reusing are increasing in general, though there is heterogeneity between different diseases. Our investigation suggested a positive correlation between data sharing as well as reuse and the research funding. Recently, Barend Mons suggested an invest 5% of research funds in ensuring data are reusable. It is irresponsible to support research but not data stewardship (Mons, 2020).
According to the analysis on data records sharing and reuse, the number of data records sharing for the following diseases may be appropriately increased in future research: Malaria, Diarrhoeal diseases and Protein-energy malnutrition. Some diseases need more funding support, such as Migraine. We believe that the reason is that scientific research is driven by both interest and demand. Compared with the amount of funding and scientific publications, the sharing and reuse of data records should be more demand-driven, which mainly comes from disease burden or unmet clinical needs. We will verify it in future research for this hypothesis. The frequently cited data records are concentrated in infectious diseases, cancer and basic health care, involving HIV infection, etc. And the cited data records come from institutions in European and American countries and regions mainly. This may be related to various openness level of health-related data in various countries and regions.
One limitation of our study using DCI is the concerns on coverage and comprehensiveness, particularly in terms of the number of repositories it indexes, although it’s increasing over time. For now, some of the citation data in the DCI are still ‘hit or miss’. And there are many directions worthy of further study. For example, the highly cited data records need to be furtherly explored to figure out how these data records are used in subsequent research, such as whether important medical evidence is formed or even written into clinical practice guidelines, so as to evaluate the value of medical data. The funding counts represent data collected from only 10 funding agencies. Similar to DCI data, which are limited to content available from the 500 or so indexed repositories, how representative and comparable are the data sources are cannot be evaluated. It may be that some diseases that are more regionally prevalent may are not as well supported by the 10 funding agencies, which could then affect the validity of the findings. We will also consider including the funding support data from more countries and international organizations to our study in the future research, e.g., using research grant information in dimensions.
References
Atal, I., Trinquart, L., Ravaud, P., & Porcher, R. (2018). A mapping of 115,000 randomized trials revealed a mismatch between research effort and health needs in non-high-income regions. Journal of Clinical Epidemiology, 98, 123–132. https://doi.org/10.1016/j.jclinepi.2018.01.006
Bai, Y., & Du, J. (2021). Measuring the impact of clinical data in terms of data citations by scientific publications. In Proceedings of the 18th International Conference on Scientometrics and Informetrics, 2021, Leuven University Press, 71–80.
Colavizza, G., Hrynaszkiewicz, I., Staden, I., Whitaker, K., & McGillivray, B. (2020). The citation advantage of linking publications to research data. PLoS ONE. https://doi.org/10.1371/journal.pone.0230416
Evans, J. A., Shim, J. M., & Ioannidis, J. P. (2014). Attention to local health burden and the global disparity of health research. PLoS ONE, 9(4), e90147. https://doi.org/10.1371/journal.pone.0090147
Force, M. M., & Robinson, N. J. (2014). Encouraging data citation and discovery with the Data Citation Index. Journal of Computer-Aided Molecular Design, 28(10), 1043–1048. https://doi.org/10.1007/s10822-014-9768-5
Huang, M., Zolnoori, M., Balls-Berry, J. E., Brockman, T. A., Patten, C. A., & Yao, L. (2019). Technological innovations in disease management: text mining US patent data from 1995 to 2017. Journal of Medical Internet Research, 21(4), 316.
Jung, Y. L., Yoo, H. S., & Kim, E. S. (2019). The relationship between government research funding and the cancer burden in South Korea: Implications for prioritising health research. Health Res Policy Syst, 17(1), 103. https://doi.org/10.1186/s12961-019-0510-6
Marshall, I. J., L’Esperance, V., Marshall, R., Thomas, J., Noel-Storr, A., Soboczenski, F., Benjamin, N., & Wallace, B. C. (2021). State of the evidence: A survey of global disparities in clinical trials. BMJ Global Health. https://doi.org/10.1136/bmjgh-2020-004145
Mons, B. (2020). Invest 5% of research funds in ensuring data are reusable. Nature, 578(7796), 491. https://doi.org/10.1038/d41586-020-00505-7
Panagopoulos, G., Tsatsaronis, G., & Varlamis, I. (2017). Detecting rising stars in dynamic collaborative networks. Journal of Informetrics, 11(1), 198–222. https://doi.org/10.1016/j.joi.2016.11.003
Park, H., & Wolfram, D. (2017). An examination of research data sharing and re-use: Implications for data citation practice. Scientometrics, 111(1), 443–461. https://doi.org/10.1007/s11192-017-2240-2
Park, H., You, S., & Wolfram, D. (2018). Informal data citation for data sharing and reuse is more common than formal data citation in biomedical fields. Journal of the Association for Information Science and Technology, 69(11), 1346–1354. https://doi.org/10.1002/asi.24049
Rubin, R. (2017). Profile: Institute for health metrics and evaluation, WA, USA. Lancet, 389(10068), 493–493. https://doi.org/10.1016/s0140-6736(17)30263-5
Shilo, S., Rossman, H., & Segal, E. (2020). Axes of a revolution: Challenges and promises of big data in healthcare. Nature Medicine, 26(1), 29–38. https://doi.org/10.1038/s41591-019-0727-5
Tenopir, C., Allard, S., Douglass, K., Aydinoglu, A. U., Wu, L., Read, E., Manoff, M., & Frame, M. (2011). Data sharing by scientists: Practices and perceptions. PLoS ONE. https://doi.org/10.1371/journal.pone.0021101
Terry Robert, F., Salm José, F., Nannei, C., & Dye, C. (2014). Creating a global observatory for health R&D. Science, 345(6202), 1302–1304. https://doi.org/10.1126/science.1258737
WHO. (2018). Global Health Estimates 2016: Disease burden by cause, age, sex, by country and by region, 2000–2016. Retrieved from Geneva.
WHO. (2019). Global observatory on Health R&D.
Yao, L., Li, Y., Ghosh, S., Evans, J. A., & Rzhetsky, A. (2015). Health ROI as a measure of misalignment of biomedical needs and resources. Nature Biotechnology, 33(8), 807–811. https://doi.org/10.1038/nbt.3276
Yegros-Yegros, A., van de Klippe, W., Abad-Garcia, M. F., & Rafols, I. (2020). Exploring why global health needs are unmet by research efforts: The potential influences of geography, industry and publication incentives. Health Research Policy and Systems, 18(1), 47. https://doi.org/10.1186/s12961-020-00560-6
Zhang, L., Zhao, W., Liu, J., Sivertsen, G., & Huang, Y. (2020). Do national funding organizations properly address the diseases with the highest burden?: Observations from China and the UK. Scientometrics, 125(2), 1733–1761. https://doi.org/10.1007/s11192-020-03572-9
Zuo, X., Chen, Y., Ohno-Machado, L., & Xu, H. (2021). How do we share data in COVID-19 research? A systematic review of COVID-19 datasets in PubMed Central Articles. Briefings in Bioinformatics, 22(2), 800–811. https://doi.org/10.1093/bib/bbaa331
Acknowledgements
The present study is an extended version of an article presented at the 18th International Conference on Scientometrics and Informetrics, KU Leuven (Belgium), 12–15 July 2021 (Bai & Du, 2021). This work was funded by the National Natural Science Foundation of China (Grant Nos. 71603280, 72074006), and the Young Elite Scientists Sponsorship Program by China Association for Science and Technology (2017QNRC001). The present study is an extended version of an article presented at the 18th International Conference on Scientometrics and Informetrics, KU Leuven (Belgium), 12–15 July 2021.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare that are relevant to the content of this article.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Bai, Y., Du, J. Measuring the impact of health research data in terms of data citations by scientific publications. Scientometrics 127, 6881–6893 (2022). https://doi.org/10.1007/s11192-022-04559-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11192-022-04559-4