Abstract
Sperm after short-term storage in vitro is widely used for artificial fertilization in aquaculture. It has been shown that short-term storage affects sperm motility characteristics, resulting in diminished fertility. However, the detrimental effects of short-term sperm storage on embryos development have remained unexplored in single-base methylome resolution. The main aim of the present study was to investigate DNA methylation in the offspring of common carp (Cyprinus carpio) derived from short-term stored sperm. Sperm were stored in artificial seminal plasma on ice (0–2 °C) for 0, 3 and 6 days in vitro, fertilization was performed using oocytes from a single female, and embryos were collected at the mid-blastula stage. In the DNA methylation study, DNA from both sperm and embryos was extracted and analysed using liquid chromatography with tandem mass spectrometry (LC–MS/MS). Concurrently, DNA methylation levels of embryos in single base were evaluated through whole genome bisulfite sequencing (WGBS). Sperm storage showed negative effects on sperm motility, viability, and DNA integrity, but had no effect on global DNA methylation of spermatozoa and resulting embryos. Results from the WGBS showed that methylation of 3313 differentially methylated regions (DMRs)-target genes was affected in the embryos fertilized with the 6-day-stored sperm, and the identified DMRs were mainly involved in cell adhesion, calcium, mitogen-activated protein kinase and adrenergic signalling, melanogenesis, metabolism and RNA transport. Such results suggest that prolongation of storage time may have certain impacts on embryonic development. These initial results provide valuable information for future consideration of the DNA methylome in embryos generated from short-term stored sperm, which are used for genetic management of broodstock in aquaculture.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Information not contained in a DNA sequence can be transmitted from parents to offspring through gametes. This is known as epigenetic inheritance, in which non-DNA sequence-based epigenetic information can be inherited across several generations (Daxinger and Whitelaw 2012; Fitz-James and Cavalli 2022). In the sequence, a cytosine base attached to a methyl group is commonly followed by a guanine base, collectively referred to as a CpG dinucleotide. DNA methylation is a widely studied heritable epigenetic modification that occurs predominantly in the CpG context in animals and plays a crucial role in regulating gene expression by recruiting proteins involved in gene repression and inhibiting transcription factors without altering the DNA sequence (Egger et al. 2004; Ohka et al. 2011). In reproduction, a number of environmental factors or physiological conditions can cause abnormal DNA methylation in fish gametes (Zhang et al. 2023a). For example, short-term and long-term storage of sperm can lead to changes in sperm DNA methylation (Cheng et al. 2021; Shazada et al. 2024; Zhang et al. 2023a). In this context, the DNA methylation of spermatozoa is strongly influenced by the use of an extender for short-term storage and the type of cryoprotectant used for cryopreservation. Cheng et al. (2023) reported that global DNA methylation changes induced by short-term-storage in common carp (Cyprinus carpio) spermatozoa can be prevented if sperm storage was carried out in an extender. Depincé et al. (2020) stated that dimethylsulfoxide and 1,2-propanediol decreased global DNA methylation in goldfish (Carassius auratus) spermatozoa, but methanol, as an effective cryoprotectant, did not allow the appearance of changes. Similarly, functions of extenders were also showed in European eel sperm (Herranz-Jusdado et al. 2019). Interestingly, epigenetic modifications in gametes under environmental and/or physiological conditions show species specificity in fishes. For instance, in contrast to goldfish sperm, global DNA methylation in zebrafish (Danio rerio) sperm increased after cryopreservation using methanol as a cryoprotectant (Depincé et al. 2020). In addition, it is worth paying attention to epigenetic reprogramming in embryos, because of its crucial role in embryonic development (Feng et al. 2010). It has been reported that sperm preserved at –20 °C without cryoprotectants is epigenetically safe to use for intracytoplasmic sperm injection, as the dynamic epigenetic reprogramming of embryos derived from frozen sperm was similar to that of embryos derived from fresh sperm (Chao et al. 2012). However, epigenetic changes in response to the environmental stressor may mediate the adaptation of organisms to the environment. In shortfin molly (Poecilia mexicana), the acclimatised epigenome under hydrogen sulfide could be inherited by subsequent generations, facilitating the adaptation of P. mexicana to sulfidic environments (Kelley et al. 2021).
In aquaculture, short-term storage of sperm is a convenient and valuable method of preserving sperm under refrigerated conditions for artificial fertilization (Cheng et al. 2022a; Shazada et al. 2024). It is most beneficial to reduce the hatchery workload on the same day that females are stripped, and to conserve male sperm if females are delayed in maturing. However, spermatozoa are unavoidably exposed to environmental stressors, such as reactive oxygen species (ROS) and natural aging during the storage (Dietrich et al. 2021). Importantly, oxidative stress in sperm may influence epigenetic reprogramming in early embryonic development (Wyck et al. 2018). Spermatozoa have compact nuclei, which distinguishes them from somatic cells. However, DNA methylation patterns and levels are changed dynamically during gametogenesis and embryonic development, it remains stable in somatic tissues. Stability of sperm DNA methylation has been reported as being critical for developing embryos (Labbé et al. 2017; Donkin and Barrès 2018). In zebrafish, the paternal DNA methylation pattern is maintained throughout early embryonic development, but the maternal genome after fertilization undergoes reprogramming to become consistent with the paternal methylation pattern (Jiang et al. 2013; Potok et al. 2013; Murphy et al. 2018).
The literature reveals that environmental factors during gamete storage may affect the epigenome (Andreu-Noguera et al. 2023) and their epigenetic reprogramming is essential for embryonic development (Potok et al. 2013; Murphy et al. 2018). Sperm storage is mostly used traditionally in artificial reproduction of economically important species in aquaculture; compared to studies in model fish species, epigenetic research is more recent and scarcer (Liu et al. 2022). Common carp is one of the major finfish species in aquaculture, with global production increasing from 1205 thousand tonnes in 1990 to 4360 thousand tonnes in 2020 (Shazada et al. 2024). Carp offspring are produced by artificial reproduction, and short-term stored or cryopreserved sperm is widely used (Shazada et al. 2024). Therefore, it is worth assessing the safety of stored sperm for the offspring in terms of their gametes’ handling/storage and their application in artificial reproduction. In our recent study, sensitive methods based on liquid chromatography with tandem mass spectrometry (LC–MS/MS) were performed to evaluate the global epigenetic level (5-mdC) in sperm and embryos. Finally, there were no significant changes in epigenetic factors (Cheng et al. 2023). Despite the absence of significant differences in global methylation levels between groups of embryos derived from fresh and stored sperm (Cheng et al. 2023), the considerable variability suggests the potential for distinct site-specific DNA methylation signatures (Khezri et al. 2019; He et al. 2022). More accurate techniques need to be developed, it can provide higher resolution at the single base level, such as whole-genome bisulfite sequencing (WGBS), which is a valuable and extensively used approach. Moreover, understanding the epigenetic regulation of different features can be helpful to guide the practical applications in aquaculture, using different methodologies. For example, the quality of common carp gametes and embryos could be enhanced in the future by epigenetic modifications of the sperm, such as epigenomic editing and epigenetic selection (Metzger and Schulte 2016; Katayama and Andou 2021; Nakamura et al. 2021).
The aim of the present study was to investigate the sperm phenotypes and DNA methylation in the common carp embryos from artificial fertilization with sperm stored for a short-term. The fresh oocytes from one female were fertilized by different stored sperm, and the differentially methylated regions (DMRs) and differentially methylated genes (DMGs) were identified in the embryos at the mid-blastula stage by direct deep sequencing to elucidate correlations between the DNA methylation profile in the embryos and the period of sperm storage.
Materials and methods
Animal ethics
The manipulations of animals in this study were performed in accordance with the relevant legal requirements and ethical standards. Specifically, the breeding and the supply of laboratory animals were authorised by the Ministry of Agriculture of the Czech Republic under reference numbers 56665/2016-MZE-17214 and 64155/2020-MZE-18134, while the use of laboratory animals was authorised under reference number 68763/2020-MZE-18134. In addition, the authors of this study, PL (CZ 03273), VR (CZ 02992), MR (CZ 01664) and OL (CZ 02815), hold a Certificate of Professional Competence according to section 15d (3) of ACT No. 246/1992 Coll. on the Protection of Animals against Cruelty, which qualifies them to perform animal experiments.
Broodstock and gamete collection
Five 3-year-old mature common carp males were chosen from broodstock with healthy and good-quality sperm. For each hormonal administration, a single intramuscular injection of carp pituitary (CP; Klatovské rybářství a.s., Czech Republic; 2 mg/kg b.w. dissolved in physiological solution) was performed to induce sperm maturation (Linhart et al. 2015). After each time of hormonal administration, the milt of each male was collected with a syringe and stored on crushed ice (0–2 °C) in vitro under aerobic conditions (see detailed description in experimental design). At first, the quality of sperm was evaluated, and the best three sperm samples with motility ~ 80% were selected for further experiments (Fig. 1). To induce final oocyte maturation, the female was treated with CP at 0.5 mg/kg b.w. (primary injection) and 2.7 mg/kg b.w. (secondary injection) with a 12 h interval. Good quality oocytes from a female (8.3 kg) in good physical condition was chosen for the fertilization test.
Experimental design and procedure for comparing fresh sperm (Fresh_SPZ) vs. stored sperm for 3 days (Store_SPZ3d) vs. stored sperm after 6 days (Store_SPZ6d) and their embryos. Three stripping procedures were performed on each male (n = 3) at 12 h post each hormonal stimulation of spermiation, and sperm samples were immediately diluted with an extender and stored on ice (0–2 °C). The sperm from the first hormonal treatment was stored for 6 days in vitro (Stored_SPZ6d), the sperm from the second hormonal treatment for 3 days (Stored_SPZ3d), and the third stripped sperm was without storage in vitro (Fresh_SPZ). In total three sperm samples were used to fertilize eggs from one female on the same day. Sperm phenotypes (sperm kinetics, membrane integrity), DNA fragmentation and global DNA methylation (5-mdC/dC (%)) were analysed. DNA methylation of embryos in the single base resolution was sequenced by WGBS. A consistent colour scheme is used throughout the figures for stored and fresh sperm and the corresponding embryos
Experimental design for sperm storage in vitro
The experiment was designed to obtain fresh sperm and stored sperm in vitro from the same males to explore the effects of sperm storage on spermatozoa epigenetics and their next generation implications in common carp (Fig. 1). After each stripping, we stored sperm of each individual male in vitro on ice to produce sperm samples with different levels of aging for artificial fertilization. Additionally, this approach allowed us to fertilize and generate offspring from the same female at the same time. Multiple hormone injections and male stripping were required to accomplish this, but it was done carefully to avoid harming the fish. In brief, three hormonal treatments were provided to the same three males at 3-day intervals (days 0, 3 and 6 of the experiment). Fresh sperm was collected at each 12 h post-hormonal treatment and immediately diluted with an extender (110 mM NaCl, 40 mM KCl, 2 mM CaCl2, 1 mM MgSO4, 20 mM Tris, pH 7.5 and 310 mOsmol/kg) (Cejko et al. 2018) at a ratio 1:1 (v:v). Fresh sperm (Fresh_SPZ), stored sperm for 3 days (Store_SPZ3d) and 6-day-stored sperm (Store_SPZ6d) were stored in vitro for 0-, 3- and 6-days post stripping (DPS), respectively, to achieve artificial fertilization at the same time. The Fresh_SPZ obtained from the third stripping was used as a control.
Evaluation of sperm phenotypes
Spermatozoa motility, velocity and viability were assessed as phenotypic traits of sperm (Cheng et al. 2023). Spermatozoa motility was activated in distilled water containing 0.25% Pluronic F-127 at room temperature (21 °C), and the percentage of motile spermatozoa and spermatozoa velocity were evaluated from recorded videos at 15 s post sperm activation using the computer-assisted sperm analysis (CASA) system as described in Cheng et al. 2022a. Spermatozoa membrane viability was assessed using the LIVE/DEAD Sperm Viability Kit (Invitrogen/Thermo Fisher Scientific Inc.). A flow cytometry with the S3e™ Cell Sorter (Bio-Rad, Hercules, CA, USA) was used to estimate the ratio of live: dead spermatozoa.
Assessment of sperm DNA integrity
The DNA integrity of sperm was evaluated using a modified alkaline single-cell gel electrophoresis (comet assay) as described previously (Gazo et al. 2022; Giassetti 2023). Briefly, microscope slides (OxiSelectST; Cell Biolabs, INC. USA) were pre-coated with 1% normal-melting-point agarose (Invitrogen; dissolved in TBE buffer (90 mM Tris base, 90 mM boric acid, 2.5 mM EDTA)) 1 day before the comet assay. A volume of 0.6 µl of sperm was diluted with 2.5 ml of PBS (phosphate buffer solution). Diluted samples (100 µl) were mixed with 800 µl of agarose (low EEO, Sigma). Finally, 40 µl of this mixture was added to the slide and allowed to solidify at 4 °C for 5 min. All slides were gently immersed in lysis buffer (2.5 M NaCl, 100 mM EDTA, 10 mM Tris, 1% Triton X-100, 10% DMSO, pH 10) and incubated overnight at 4 °C. After the cells were lysed, the buffer was removed, and the slides were washed in TBE, and placed in a gel tank filled with electrophoresis buffer (TBE). Electrophoresis was performed for 15 min at 50 V. The slides were washed three times with distilled H2O following electrophoresis. The washed slides were dehydrated with 70% ethanol, air-dried, and then kept at 4 °C.
For agarose rehydration and DNA staining, 50 μl of PBS containing SYBR Green Staining Solution (Invitrogen) was added to the slides. The slides were analysed using an Olympus BX 50 fluorescence microscope at 400×. A minimum of 30 images were captured per sample. Comet Assay Software Project (CASP) software (version 1.2.3) was used to analyse the images.
Artificial fertilization in vitro
To eliminate female impacts on paternal transgenerational inheritance, only one female was involved in the fertilization test. To ensure successful fertilization and meet the requirements for embryo sampling, 5 g of eggs (~ 4000 eggs) were fertilized at a ratio of 500,000 spermatozoa per egg (Linhart et al. 2015) with 10 ml of activation solution (45 mM NaCl, 5 mM KCl, 30 mM Tris, pH 8.0, 160 mOsmol/kg) (Perchec et al. 1996). Prior to the fertilization experiment, spermatozoa concentration was measured using a Bürker cell hemocytometer (Marienfeld, Germany) with a final dilution of 5000-fold. Fertilized eggs were incubated in Petri dishes at 21 °C. The embryo samples at the mid-blastula stages were collected at 7.5 h post fertilization (HPF). Fertilization, hatching and malformation rates were calculated using the formula: fertilization rate (%) = fertilized eggs/total eggs *100, hatching rate (%) = hatched embryos/total eggs *100, and malformation ratio (%) = abnormal larvae/total hatched larvae *100.
Sampling and DNA extraction
The DNA methylation level was assessed in fresh and stored sperm and in samples of mid-blastula embryos. In detail, a 1.8 ml cryotube containing 2 μl of each sperm sample was snap‐frozen in liquid nitrogen and kept at − 80 ℃ until further use. The DNA isolation kit (Qiagen DNeasy, Hilden, Germany) was used to isolate sperm genomic DNA according to the manufacture’s protocol. The developed mid-blastula embryos (40–60 embryos/tube) from each treatment were sampled at 7.5 HPF. Before sampling, embryos were washed in triplicate with RNase- and DNase-free molecular biology-grade water (Invitrogen). DNA from embryo samples was extracted using a modified SDS-based DNA extraction method according to Natarajan et al. (2016). The DNA concentration was determined using a Nanophotometer Pearl (Implen, Munich, Germany). Finally, the gDNA's purity and integrity were evaluated by electrophoretic separation and visual assessment in a 1.5% agarose gel.
Assessment of global DNA methylation using isotope-dilution LC–MS/MS
Ten μg of DNA from each group of sperm and embryo samples were hydrolysed to 2’-deoxynucleosides using micrococcal nuclease from Staphylococcus aureus, bovine spleen phosphodiesterase and calf intestinal alkaline phosphatase (all from Sigma-Aldrich, Taufkirchen, Germany) as described previously (Schumacher et al. 2013). dC-[15N2,13C1], and 5-mdC-d3 (both from Toronto Research Chemicals, Toronto, Canada) were used for quantification of dC and 5-mdC in DNA hydrolysates by isotope-dilution LC–MS/MS as reported recently (Gerecke et al. 2018). In brief, nucleoside analytes were separated using a Poroshell 120 EC-C8 column (3.0 × 150 mm, 2.7 µm) fitted in a 1290 Infinity II HPLC linked to an Ultivo (G6465) triple-quadruple mass spectrometer (all from Agilent Technologies, Waldbronn, Germany). Electrospray ionization (ESI+) was used to create protonated precursor ions, and the MS/MS detector was set to multiple reaction monitoring (MRM). For quantification, the following mass transitions were used (qualifier mass transitions are in parentheses): dC: m/z 228.1 → 112.0 (94.9); [15N2,13C1] dC: m/z 231.1 → 114.9 (97.9); 5-mdC: m/z 242.1 → 126.0 (109.1), and 5-mdC-d3: m/z 245.1 → 129.0 (112.0). Using the MassHunter program (Quantitative Analysis version 10.1) from Agilent Technologies, peak regions were assessed, and nucleoside concentrations in the DNA hydrolysates were computed based on the quantities of internal standards utilized.
DNA library construction, whole-genome bisulfite sequencing, data quality control and mapping
Over 30 µg of total DNA were sent from each embryos sample (fertilized with three males) to Admera Health Biopharma Services for WGBS analysis. In detail, isolated genomic DNA was quantified using the Qubit 2.0 DNA HS Assay (Thermo Fisher Scientific, Massachusetts, USA) and the quality was assessed by TapeStation genomic DNA Assay (Agilent Technologies, California, USA). Then, following the manufacturer’s protocol, bisulfite conversion was performed using the EZ DNA Methylation-Gold Kit (Zymo Research, California, USA), and DNA was fragmented to 300–400 bp. After the bisulfite conversion, library preparation was performed using Accel-NGS® Methyl-Seq DNA Library kit (Swift Biosciences, Michigan, USA). Thereafter, library yields were determined using the Qubit 2.0 DNA HS Assay and the quality of the libraries generated was assessed using the TapeStation HSD1000 ScreenTape (Agilent Technologies Inc., California, USA). The final libraries quantity was assessed by APA SYBR® FAST qPCR with QuantStudio® 5 System (Applied Biosystems, California, USA). Based on qPCR QC values, equimolar pooling of libraries was performed. A 20% PhiX Spike In was added to the samples to provide a bisulfite conversion efficiency and added during the sequencing to ensure sequencing quality. Finally, samples were sequenced on an Illumina® NovaSeq S4 (Illumina, California, USA) with a read length configuration of 150 PE to receive 188 M reads per sample (94 M in each direction).
We conducted fundamental statistical analyses on the read quality of the raw data using FastQC (fastqc_v0.11.5). Thereafter, the read sequences generated by the Illumina pipeline in FASTQ format were then pre-processed using Trimmomatic software (Trimmomatic-0.36) with the parameter (SLIDINGWINDOW: 4:15; LEADING:3, TRAILING:3; ILLUMINACLIP: adapter.fa: 2: 30: 10; MINLEN:36). After filtering the reads, the remaining reads were considered as clean reads, and all subsequent analyses were conducted using this set of reads. Finally, upon completion of data cleaning, we utilized FastQC to generate basic statistical metrics that assessed the quality of the resulting reads.
Clean data were obtained after filtering and then the Bismark software (version 0.16.3; Krueger and Andrews 2011) was used to perform alignments of bisulfite-treated reads to a reference genome (assembly accession: GCF_000951615.1). The reference genome was firstly transformed into bisulfite-converted version (C-to-T and G-to-A converted) and then indexed using bowtie2 (Langmead and Salzberg 2012). Before being directly matched to similarly converted versions of the genome, sequence reads were also transformed into fully bisulfite-converted versions (C-to-T and G-to-A converted). The methylation status of every cytosine site in the sequence reads that result in a distinct best alignment from the two alignment procedures (original top and bottom strand) is then inferred by comparing them to the typical genomic sequence. The same reads aligned to the same regions of the genome were considered duplicated. Sequencing depth and coverage were summarised using deduplicated reads.
Analysis of methylation levels and identification of DMRs
To identify the methylation site, we modelled the sum Mc of methylated counts as a binomial (Bin) random variable with the methylation rate. After the methylation sites were determined, the methylation level (ML) for each window or C site was calculated according to the formula ML = reads (mC)/reads (mC) + reads (umC). The motif of different sequencing contexts CpG, CHG and CHH (where H is A, C or T) was determined to reveal the sequence characteristics at the upstream and downstream regions of methylated cytosine (C) in the different context sequences. DMRs were identified using DSS software (parameter: smoothing = TRUE, smoothing.span = 200, delta = 0, p.threshold = 1e-05, minlen = 50, minCpG = 3, dis.merge = 100, and pct.sig = 0.5) (Park and Wu 2016). In detail, DMRs were classified as DMRs if they had a minimum length of 50 bp, contained at least three CpG sites, and were at least 50% statistically significant differentially methylated CpGs (p < 1e−05). The DMRs were ranked by the sum of the test statistics of the contained CpG sites. We determined the DMGs whose gene body region from transcriptional start sites (TSS) to transcription end sites (TES) or the promoter region (upstream 2 kb from the TSS) overlapped with the DMRs in accordance with the distribution of DMRs in the genome.
The term "differentially methylated genes" (DMGs) was used to describe whose promoter or gene body regions (upstream 2 kb from the transcription start sites, or TSS) overlapped the DMRs. Additionally, the methylation levels of genomic functional regions were also analysed. These regions included promoter, exon, intron, CGI (CpG island) and CGI shore (defined as the 2 kb sequence flanking a CGI). The average rate of methylation was calculated for each functional genomic region.
Gene Ontology (GO) enrichment analysis of genes associated with DMRs was performed using the GOseq R package (Young et al. 2010). GO terms with a corrected P value of less than 0.05 were considered significantly enriched to be DMR-related. We conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis to identify significantly enriched metabolic or signalling pathways in genes with DMRs compared to the whole genome background. The statistical enrichment of DMR-related genes in the KEGG pathway was examined using the KOBAS program (Mao et al. 2005).
Statistical analysis
The normality of the distribution and homogeneity of variances for all data were checked using Shapiro–Wilk and Levene’s tests. Statistical significance of sperm phenotypic parameters, mdC/dC (%) from LC–MS/MS was tested either using one-way ANOVA followed by a Tukey test or the nonparametric Kruskal–Wallis test followed by a Dunn pairwise comparison. All statistical analyses were performed with GraphPad Prism 9.1.0 (GraphPad Software, San Diego, CA, USA) and RStudio version 3.6.3. Differences were considered significant at p < 0.05 except for special instructions.
Results
Short-term storage decreases sperm motility traits, DNA integrity and fertilizing ability
Comparison of biological traits of sperm between fresh and short-term stored sperm showed significant decreases in percentage of motility (Fig. 2A) and increase in percentage of sperm with DNA fragmentation (Fig. 2D) after 3 or 6 DPS (p < 0.05). However, a non-significant trend towards a decrease was observed for curvilinear velocity (VCL) (Fig. 2B), and a reduction in viability after 6 DPS (Fig. 2C). The fertilization and hatching rates were decreased when 6-day-stored sperm was used for fertilization (p < 0.05; Fig. 2F). Due to a high sperm: egg ratio used in the fertilization experiment, the fertilization and hatching rates were also high for short-term stored sperm (> 90%). Meanwhile, no changes in larval malformation were observed (p > 0.05; Fig. 2F). These results showed that storage-induced decreases in sperm quality was accompanied by fertility loss, although it did not have an apparent effect on embryonic development to hatching.
Effects of short-term storage on sperm quality, fertility and global DNA methylation. A Percent of spermatozoa motility; B curvilinear velocity (VCL) of motile spermatozoa; C percentage of live spermatozoa with intact membrane; D DNA fragmentation levels (defined by the percent of tail DNA); E fertilization and hatching rates of short-term stored sperm; F malformation rate in the embryos; G, H global DNA methylation [5-mdC/dC (%)] of sperm and embryos at mid-blastula; I average methylation level of all the genome cytosine sites of mid-blastula; J the average methylation level of all the genome CpG-context cytosine sites of mid-blastula. Statistical significance was determined using one-way ANOVA followed by a Tukey test; data of sperm, embryos and fertilization from each group (control group: Fresh_SPZ; 3-day-sperm storage group: Store_SPZ3d and 6-day-sperm storage group: Store_SPZ6d) represent the mean ± S.D. of three biological replicates (*p < 0.05; ***p < 0.001; ****p < 0.0001)
Short-term storage does not alter global DNA methylation in sperm and embryos
The 5-mdC/dC in short-term stored sperm and their embryos remained unchanged (p > 0.05; Fig. 2E, G). Also, mean mC and mCpG in the embryos derived from short-term stored sperm were similar to those derived from fresh sperm (p > 0.05; Fig. 2H, I). Taken together, these results indicated that short-term storage decreased sperm quality, but had no significant effects on global DNA methylation of the derived embryos.
Data generation and mapping to the reference genome
To reveal the differences in the DNA methylation at the whole genome level of the mid-blastula embryos derived from fresh and short-term stored sperm, we performed WGBS of embryos with a mean yield of 188 million reads. After quality control, 48–58% of the reads were uniquely mapped to the reference genome, and an average of 90% base covered ≥ 5 × . The coverage depths in the embryos from fresh and stored sperm were at least above 7.5 (Supplementary Table S1).
DNA methylation in the sequencing contexts
The specific DNA methylation level of the three sequence contexts, CpG, CHG and CHH, was analysed for the embryos derived from fresh and stored sperm samples. The average methylation levels at C-sites in the embryos originating from short-term stored sperm were similar; 7.95, 7.86 and 7.93% for fresh (Fresh_SPZ), 3-day-stored sperm (Store_SPZ3d) and 6-day-stored sperm (Store_SPZ6d), respectively (Fig. 2H and Table 1). The mean methylation levels of CpG were higher than CHG, and CHH in the embryos that originated from fresh or short-term stored sperm. The values were 79.73, 0.93 and 1.01% in the embryos obtained from fresh sperm, 79.65, 0.78 and 0.86% in the embryos obtained from 3-day-stored sperm, 79.74, and 0.90 and 1.00% in the embryos obtained from 6-day-stored sperm (Table 1). Since most mC sites covered by CpG contexts, only DMRs in CpG contexts were considered for subsequent analyses. The methylation level in the CpG context did not differ among the embryos obtained from short-term stored sperm (p > 0.05; Fig. 2I). The CpG methylation on each chromosome in the embryos from stored sperm varied from that in the embryos originating from the fresh sperm (Supplementary Fig. S1).
According to cluster analysis based on CpG methylation levels, the samples were distributed across three clusters, each containing two, three and four samples. However, the samples of various stored sperm groups did not appear to cluster together (Fig. 3A). Meanwhile, a heat map of DNA methylation based on the same criteria showed a very high correlation between the treatments within the same male (Pearson’s correlation coefficient > 0.94) (Fig. 3B).
Clustering and correlation of analysis of samples based on CpG methylation level. A Hierarchical clustering by CpG methylation levels in common carp embryos obtained from fresh and short-term stored sperm; B heat map and correlation analysis based on CpG data among common carp embryos obtained from fresh and short-term stored sperm. Numbers in each cell represent the pairwise Pearson’s correlation scores. Data obtained from sperm of three males with different storage time: fresh, 3-days- and 6-days-stored sperm from male 1 (Fresh_SPZ1, Stored_SPZ3d1, Stored_SPZ6d1), male 2 (Fresh_SPZ2, Stored_SPZ3d2, Stored_SPZ6d2), and male 3 (Fresh_SPZ3, Stored_SPZ3d3, Stored_SPZ6d3)
DNA methylation in the genomic elements
DNA methylation levels in different genomic elements were analysed in the embryos produced from fresh and short-term stored sperm (Fig. 4). Generally, the average CpG methylation levels were high in the CpG island and CGI shore and low in the 5’ untranslated region (5’ UTR) and promoter regions (Fig. 4A-C). Comparison between embryos showed that (a) the average methylation levels in several genomic regions (such as CpG island, CGI shore, promoter and 5’ UTR regions) were slightly higher in the embryos from fresh sperm than those of 3-day-stored sperm (Fig. 4A), (b) the average methylation levels in genomic regions of CpG island were higher (although not in the promoter region) in the embryos of 3-day-stored sperm than those of 6-day-stored sperm (Fig. 4B), and (c) the average methylation levels showed similar trends in the embryos of fresh sperm vs. those of 3-day-stored sperm (Fig. 4C). Analyses of DNA methylation levels in the genome upstream, gene body and downstream showed lower CpG methylation at upstream and downstream regions compared to that in the gene body region (Fig. 4D-F). In this context, the CpG methylation levels in all these regions in the fresh sperm were similar as those in the 3-day-stored sperm (Fig. 4D). Also, the CpG methylation levels of upstream and downstream regions in the embryos of fresh sperm and 3-day-stored sperm were lower than embryos of 6-days-stored sperm (Fig. 4E, F).
Comparison of methylation levels on different genomic elements and distribution of up-stream and down-stream among embryos derived from fresh sperm (Fresh_SPZ), stored sperm for 3 days (Store_SPZ3d) and for 6 days (Store_SPZ6d) in vitro. A–C Density plot of methylation levels on different genomic elements in CpG; D-F density plot of methylation levels on different regions of up-stream, gene body and down-stream. (n = 3 for each group)
DMRs among embryos developed from fresh and stored sperm
In total, 4761 DMRs with annotation were identified between the embryos of fresh and embryos of 3-days-stored sperm, including 2734 hypo-DMRs in 3-days-stored sperm and 2027 hyper-DMRs. There were 8624 DMRs between embryos of 3-day-stored sperm and embryos of 6-day-stored sperm (4069 hypo-DMRs and 4555 hyper-DMRs); 7726 DMRs between embryos of fresh and 6-day-stored sperm (4062 hypo-DMRs and 3664 hyper-DMRs) (Supplementary Fig. S2). The average DMR methylation level and distribution pattern were similar between embryos of fresh sperm and 3-day-stored sperm (Fig. 5A). However, there were more hyper- and hypo methylation regions in the embryos of 6-day-stored sperm than those obtained from 3-day-stored sperm (Fig. 5B, C).
Analysis of differential methylation levels and regions (DMRs). A–C Violin plots of methylation levels of DMRs in the embryos original from fresh sperm (Fresh_SPZ), 3-day-stored sperm (Store_SPZ3d) and 6-day-stored (Store_SPZ6d) in vitro. All comparisons: P < 0.0001. D–F Venn diagrams of DMR-anchored genes in gene region including all types, hyper DMR and hypo DMR-target genes in CpG content of embryos originating from fresh sperm (Fresh_SPZ), 3-day-stored sperm (Store_SPZ3d) and 6-day-stored sperm (Store_SPZ6d) in vitro. G–I Venn diagrams of DMR promoter-target genes including all types, hyper DMR and hypo DMR-target genes in CpG content of embryos original from fresh sperm (Fresh_SPZ), 3-day-stored sperm (Store_SPZ3d) and 6-day-stored sperm (Store_SPZ6d)
In the embryos of 3-day-stored and 6-day-stored sperm at all different gene regions (CGI, CGI shore, promoter, TSS, 5’ UTR, exon, intron, 3’ UTR and TES), the number of hyper-methylated DMRs was also significantly higher than hypo-methylated DMRs (Supplementary Fig. S3A, C). However, an opposite trend was observed between 3-day- and 6-day-stored sperm (Supplementary Fig. S3B). By anchoring the DMRs to the gene body (from TSS to TES) and promoter region, 2053, 3687 and 3313 differentially methylated genes (DMGs) in embryos from fresh vs. 3-days-stored sperm, 3-days-stored sperm vs. 6-days-stored sperm, and fresh vs. 6-days-stored sperm, respectively, were identified from CpG-type DMRs (Fig. 5D–I).
A total of 1516, 2711 and 2451 differentially methylated genes (DMGs) in the gene body was identified between embryos from fresh sperm vs. 3-day-stored sperm, 3-day-stored sperm vs. 6-day-stored sperm, and fresh sperm vs. 6-day-stored sperm (Fig. 5D), including 904, 1386 and 1373 upregulated DMGs (Fig. 5E) and 722, 1561 and 1261 downregulated DMGs, respectively (Fig. 5F). Moreover, 321 and 206 common DMGs were either upregulated or downregulated in the embryos of fresh sperm compared to those of 3-day-stored and 6-day-stored sperm (Fig. 5E, F). In the promoter regions, more common DMGs (56) were identified in downregulated genes than upregulated genes (11) in the above comparisons (Fig. 5G–I).
Functional enrichment analysis for genes with differentially methylated genes (DMGs)
To reveal the biological functions of differentially methylated genes among embryos derived from fresh and stored sperm, we performed pathway analysis of differentially methylated genes based on the GO and KEGG database. Five GO terms were notably enriched from GO enrichment analysis, and the results indicated that promoter hypo-DMGs in the embryos only from sperm groups of fresh vs. 6-days-stored were enriched in the biological process related to cell–cell adhesion and homophilic cell adhesion (Table 2, Supplementary Table S2). However, in the analysis of promoter hyper-DMGs, hyper- and hypo-DMGs in the gene body, there were no significant differences in the genes of the biological process (BP), cellular component (CC) and molecular function (MF) classifications among three groups (p > 0.05; Supplementary Fig. S4 and S5).
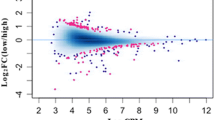
In addition, KEGG enrichment analysis showed only oxidative phosphorylation was significantly enriched in promoter hyper-DMGs between fresh and 3-day-stored sperm groups, and hypo-DMGs when compared with 3-day-stored and 6-day-stored sperm groups (Supplementary Table S3). The gene body hyper-DMGs were enriched in KEGG pathways, such as neomycin, kanamycin and gentamicin biosynthesis, calcium signalling pathway, adrenergic signalling in cardiomyocytes, regulation of actin cytoskeleton and melanogenesis between fresh and 6-day-stored sperm (Fig. 6C) and pathways related to focal adhesion, cell adhesion molecules (CAMs) and RNA transport enriched in gene body hyper-DMGs between 3-day-stored and 6-day-stored sperm (Fig. 6B). In the gene body hypo-DMGs, pathways included but were not limited to starch and sucrose metabolism, neomycin, kanamycin and gentamicin biosynthesis, galactose metabolism, Apelin signalling pathway and RNA transport (fresh sperm vs. 3-day-stored sperm; Fig. 6D); MAPK signalling pathway and Adherens junction (fresh sperm vs. 6-day-stored sperm; Fig. 6E). Only three pathways of cardiac muscle contraction, cell adhesion molecules (CAMs) and adrenergic signalling in cardiomyocytes were different between 3-day-stored sperm and 6-day-stored sperm in vitro (Fig. 6F). Detailed information on the KEGG enrichment analysis is shown in supplementary table S3.
A–C KEGG enrichment analyses of DMGs in gene body hyper-DMGs and D–F hypo-DMGs related pathways with the most significant P value of different methylation genes among embryos derived from fresh sperm (Fresh_SPZ), 3-day-stored sperm (Store_SPZ3d) and 6-day-stored sperm (Store_SPZ6d) in vitro. The x-axis indicates percentages of differential methylated genes belonging to the corresponding pathway. The sizes of bubble represent the number of DMGs in the corresponding pathway, and the colours of the bubble represent the enrichment P-value of the corresponding pathway. Red colour shows the significant pathways with corrected p-value < 0.05
Discussion
DNA methylation is one of the most important epigenetic modifications involved in regulating growth, reproduction, disease resistance and stress response in aquaculture species (Liu et al. 2022). However, little is known about its function when short-term stored sperm is used for artificial fertilization in hatcheries. Fish sperm, like those of other vertebrates (Darszon et al. 1999), are immobile in the sperm duct and spermatozoa can be stored in the natural or artificial seminal plasma in a quiescent state for a short time without loss of motility and fertilizing capacity (Shazada et al. 2024). In the present study, the main objective was to investigate DNA methylation profiles of embryos produced from short-term stored sperm in common carp using WGBS. We also assessed sperm functions after the short-term storage. Taken together, sperm motility characteristics, DNA integrity and viability were reduced, but global DNA methylation of spermatozoa and resulting embryos were not affected.
Sperm motility is an essential phenotype to manifest fertilization capacity and pathway to offspring formation. In this study, the method of sperm storage was similar to that used in our previous studies (Cheng et al. 2022a, b; Zhang et al. 2023b). Sperm motility traits were measured immediately after storage, and results were consistent with our recent study (Zhang et al. 2023b), which indicated that the percentages of motile spermatozoa and spermatozoa viability were significantly decreased after storage at 6 DPS even with elevated temperature after sperm storage. No significant decrease in spermatozoa velocity was observed after 6 DPS compared to fresh sperm, which is consistent with that of the fertilization rate. This is due to the fertilization rate being primarily dependent on sperm velocity (Linhart et al. 2005). However, the storage procedure must be reliable in order to preserve the genetic and epigenetic contents of sperm during the storage period. In terms of sperm DNA integrity, there was an increased level of DNA fragmentation in 6-day-stored sperm with respect to the tail DNA (Fig. 2). This suggests that short-term storage causes damage to sperm DNA in common carp, which is consistent with previous studies in mammals and fishes (Pérez-Cerezales et al. 2009; Shaliutina et al. 2013; Khezri et al. 2019). However, the stability of sperm chromatin shows species specificity during in vitro storage, for example, DNA damage was increased in Russian sturgeon (Acipenser gueldenstaedtii), Siberian sturgeon (Acipenser baerii) and rainbow trout (Oncorhynchus mykiss) sperm at 6-, 3- and 2-days post-storage, respectively (Pérez-Cerezales et al. 2009; Shaliutina et al. 2013). DNA fragmentation and sperm membrane integrity have been reported to be associated with oxidative stress (Gazo et al. 2015). In addition, in terms of global DNA methylation, the current result is consistent with a previous study (Cheng et al. 2023), which suggested that sperm preservation in extenders can, to a large extent, mitigate the adverse effects of ROS and safeguard the cell structure.
In general, the contexts of DNA methylation have been identified as CpG, CHH and CHG, but in animal genomes, DNA methylation mostly occurs on the cytosines of a CpG dinucleotide (Lee et al. 2010). In fishes, high levels of CpG methylation have been reported, such as 94% in zebrafish (Potok et al. 2013), 98% in grass carp (Ctenopharyngodon idellus) (He et al. 2022), 71–73% in Chinese perch (Siniperca chuatsi) (Pan et al. 2021), 98% in tiger pufferfish (Takifugu rubripes) (Zhou et al. 2019), and 76–78% in ricefield eel (Monopterus albus) (Chen et al. 2021). In this study, the average global methylation level in a CpG context across the nine different common carp embryo samples was 79.71% (Table 2), and lower methylation levels were observed in the CHH (0.86–1.01) and CHG (0.78–0.93). It was similar to our previous study which showed 79% mCpG in common carp spermatozoa (Cheng et al. 2021). Generally, DNA methylation levels vary depending on the specific tissue, sex and stage of development (Strömqvist et al. 2010; Wang and Bhandari 2019). Based on the changes in sperm DNA fragmentation and from a previous study in sperm DNA methylome (Cheng et al. 2021), the question is raised of whether short-term induced damage to sperm DNA affects offspring development. Clustering analysis and Pearson correlation based on CpG methylation levels showed no distinct clustering and a very high positive correlation between males (Fig. 3). These findings show slight variation between the treated samples and imply that the specific differences in DNA methylation by sperm storage for 6 days are small in common carp embryos. Since the embryo samples from the different stored sperm belonging to the same male displayed similar methylation levels, genetic diversities may have a greater effect on DNA methylation than gamete manipulation.
Analysis of DMRs revealed a considerable increase in the number of hyper/hypo DMRs and DMRs target genes in the Fresh_SPZ vs. Store_SPZ6d and Store_SPZ3d vs. Store_SPZ6d comparison compared to the Fresh_SPZ vs. Store_SPZ3d comparison (Fig. 5). The association between sperm manipulation and sperm DNA methylation is inconsistent in the literature. Most of the studies have investigated long-term (cryopreserved) stored sperm. For example, cryopreservation has been shown to have effects on epigenetic markers that vary among animals (Chatterjee et al. 2017), cryopreservation agents (Depincé et al. 2020), and accuracy of the methods used in the investigation (Kurdyukov and Bullock 2016). Some studies have also shown that the influence of short-term storage of sperm on DNA methylation is due to the different levels of DNA fragmentation, and there is an increase in the number of downstream genes and their associated regulatory pathways that may be impacted with increasing DNA fragmentation in spermatozoa (Khezri et al. 2019). Sperm DNA fragmentation significantly correlates with fertility performance and abnormal embryo development (Gosálvez et al. 2014). On the other hand, the sperm methylome is already determined during spermatogenesis (Ben Maamar et al. 2022); but there are factors that can mutate the DNA methylome of sperm, including the DMRs target genes. For example, there is evidence showing that oxidative stress produced during storage leads to changes in DNA methylation patterns in spermatozoa (Khezri et al. 2019). Oxidative stress not only leads to DNA fragmentation but also causes damage to sperm membranes and mitochondria, resulting in sperm with abnormal morphology and flagella. Consequently, sperm motility and the number of normal spermatozoa decreases. The higher frequency of DNA fragmentation observed in spermatozoa after 3 and 6 days of storage in this study may be due to possible elevated levels of oxidative stress (Fig. 2E).
The regional analysis showed that most of the DMRs were annotated to be present in the intron, CpG islands (CGI) and CGI shore regions (Fig. S3). Methylation in gene promoters has drawn the greatest attention in the conventional model because it is frequently linked to transcriptional silence. However, recent research indicates that in some specific genes, the first intron's methylation is related to gene expression (Anastasiadi et al. 2018). In accordance with the present results, the methylation pattern was similar to previous studies on Chinese perch, which showed the most DMR was concentrated in the intronic regions (Pan et al. 2021). Meanwhile, intronic enhancers that interact with the promoters of the relevant genes could partially explain it. Additionally, CGIs are short-interspersed DNA sequences with a higher CpG frequency than other regions in the genome. At the same time, CGI shores (up to 2 kb distant) are the immediate regions flanking CGIs (Deaton and Bird 2011). CGI methylation is frequently linked to transcriptional silence because CGIs are typically found near transcription start sites (Jones 2012). In the present study, embryos obtained from fresh sperm had slightly higher methylation levels than those obtained from stored sperm (Fig. 3), suggesting that stored sperm induce some genes transcription activity during embryogenesis.
By overlapping the differentially methylated regions and functional gene regions, 378 differentially methylated genes were identified among three embryo groups in this study. In addition, 1373 significantly hypo-DMGs were identified in the methylome analysis. GO enrichment analysis identified the biological process of differentially methylated genes between the embryos produced by fresh and stored sperm. These DMGs were involved in GO terms of cell adhesion. The cell adhesion molecules (CAMs) pathway was also enriched in KEGG analysis. Cell–cell adhesion molecules play crucial roles, especially in the early stages of reproduction, such as gamete transport, sperm-oocyte interaction, embryonic development and implantation (D’Occhio et al. 2020). The results from the DMGs suggest that prolongation of storage time may have higher impact on embryonic development. In general, most external fertilization such as fish has a lower selective pressure than mammals, raising the chances of successful fertilization with spermatozoa that have DNA damage (Pérez-Cerezales et al. 2010; Herráez et al. 2017). In rainbow trout embryos, genes belonging to cell adhesion are important in vascular development (Vehniäinen et al. 2016). Notably, studies have demonstrated that fertilization with spermatozoa exhibiting oxidative stress is associated with aberrant early cleavage events and changes in embryo transcript abundance. Mammalian embryos produced from ROS-treated sperm were identified with a similar pathway (cell adhesion) associated with differentially expressed genes (Burruel et al. 2013). Sperm preservation in extenders may also directly affect the epigenome of the paternal DNA. For instance, a recent study has shown that boar (Sus scrofa) sperm after preservation display various increases in the DNA fragmentation index (DFI) among individuals, and genes corresponding to low–high DFI comparison were associated with crucial processes such as membrane function, metabolic cascade and antioxidant defence system (Khezri et al. 2019). In the present study, it is possible that 6-days-stored sperm underwent oxidative stress according to an increase in the DNA fragmentation level (Fig. 2E). This may be the reason behind the observed induced changes in the epigenome of embryos resulting from 6-days-stored sperm.
Studies associated with the DNA methylome and transcriptome of the sperm following short-term storage and their sired offspring are few compared to cryopreserved sperm. The transcriptome and methylome analyses of cryopreserved sperm of viviparous black rockfish (Sebastes schlegelii) revealed that the cryopreservation affected the expression of 179 genes and methylation of 1266 genes in spermatozoa. These genes associated with differential expression or methylation were highly enriched in KEGG pathways of phospholipase D signalling pathways and xenobiotic and carbohydrate metabolism pathways, and GO terms of death, G-protein coupled receptor signalling pathways, and response to external stimulus (Niu et al. 2022). Fertilization with frozen-thawed sperm in mammals downregulates genes relevant to early embryo development, such as in biological pathways related to oxidative phosphorylation, DNA binding, DNA replication and immune response (Ortiz-Rodriguez et al. 2018). In the case of short-term stored sperm, it has been observed that transcripts of genes involved in energy metabolism and stress response were decreased gradually (Yang et al. 2021). These findings suggest that disruption to the transcriptome and DNA methylome of preserved sperm may have an adverse impact on the development of embryos.
In our previous study, more than a thousand of DMRs (875 methylated and 854 unmethylated) were found between fresh and 96 h-stored common carp sperm (Cheng et al. 2021). In the present study, under the protection function of extenders, various KEGG pathways were identified from embryos obtained with 3-day-stored (72 h) and 6-day-stored (144 h) sperm, and higher changes in genes related to DMRs were identified following 6 days of storage than 3 days of storage. For example, at 6 days of post storage, KEGG pathways suggested short-term storage influenced calcium signalling, adrenergic signalling in cardiomyocytes, regulation of actin cytoskeleton, and melanogenesis in the resulting embryos. However, 3 days of sperm storage changed the pathways involved in metabolism, Apelin signalling and RNA transport, which play an irreplaceable role in the regulation of the heart, lymphatic development and precise spatiotemporal control of gene expression. Taken together, these results suggest that short-term storage of sperm may affect embryonic development in these aspects. For example, Fang et al. (2016) showed that DNA methylation played a crucial role in embryonic cardiomyocytes because the knockdown of DNA methyltransferase 3a was found to alter gene expression and inhibit the function of embryonic cardiomyocytes. Additionally, the crucial and varied roles performed by actin regulation during morphogenesis, including signals regulating by the Rho Family of Small GTPases and MAP Kinase Cascades have been found (Woolner and Martin 2006). Similar pathways in sperm after treatment with ROS have previously indicated changes in transcript abundance for genes related to actin cytoskeleton organization compared to good sperm (Burruel et al. 2013). In addition, melanogenesis is the process by which melanin, a pigment responsible for coloration, is produced and deposited in various cells and tissues. The functions of melanogenesis in fish embryos may involve camouflage, UV protection, temperature regulation, immune defence, oxygen balance and cell signalling and development. Sperm itself cannot directly regulate melanin production in embryos, but sperm provides paternal genetic information, including genes related to melanin production. These genes can be expressed when an embryo is developing, and they take a role in controlling the routes and procedures involved in the production of melanin. However, these results are insufficient to indicate the influence of stored sperm on the resulting embryos. Multiple factors from sperm, such as histone modifications and noncoding RNA, determine the gene expression and protein synthesis in embryos. Thus, the systemic examination of the epigenetic status and other multi-omics simultaneously is warranted.
Conclusion
The current work provides evidence that sperm short-term storage may impact DNA methylation alterations in the resulting embryos. Prolongation of storage duration shows higher impacts on the DNA methylation of the derived embryos. Furthermore, DMR analysis revealed that genes involved in cell adhesion, calcium signalling, mitogen-activated protein kinase (MAPK) and adrenergic signalling, metabolism, and RNA transport were primarily affected in embryos derived from sperm stored for 6 days in vitro.
This is the first investigation elaborating on the single-base DNA methylome as an epigenetic alteration in embryos originating from the short-term stored sperm samples in common carp. Using common carp as a fish model organism provided us with an advantage to perform multi-stripping of a male and to collect enough volume of sperm to analyse sperm motility, DNA integrity, viability, epigenetics and to conduct fertility tests together. The current study reports enough robust data from the 180 mid-blastula stage embryos in each group of fresh sperm, and sperm stored for 3 and 6 days in vitro obtained from the same male's sperm samples under either fresh or stored conditions. Our results provide valuable information on genetic management of broodstock of common carp in aquaculture. However, the parallel transcriptomic analysis in future research would be informative and may reveal further consequential epigenetic changes in the resulting progeny from the short- or long-term stored gametes.
Availability of data and materials
Data will be made available on request.
Abbreviations
- LC–MS/MS:
-
Liquid chromatography with tandem mass pectrometry
- WGBS:
-
Whole-genome bisulfite sequencing
- DPS:
-
Days post stripping
- CASA:
-
Computer-assisted sperm analysis
- DMRs:
-
Differentially methylated regions
- DMG:
-
Differentially methylated genes
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopaedia of genes and genomes
- Fresh_SPZ:
-
Fresh sperm
- Store_SPZ3d:
-
In vitro Stored sperm for 3 days
- Store_SPZ6d:
-
In vitro Stored sperm for 6 days
- TSS:
-
Transcriptional start sites
- TES:
-
Transcription end sites
- 5’ UTR:
-
5’ Untranslated region
References
Anastasiadi D, Esteve-Codina A, Piferrer F (2018) Consistent inverse correlation between DNA methylation of the first intron and gene expression across tissues and species. Epigenetics Chromatin 11:37. https://doi.org/10.1186/s13072-018-0205-1
Andreu-Noguera J, López-Botella A, Sáez-Espinosa P, Gómez-Torres MJ (2023) Epigenetics role in spermatozoa function: Implications in health and evolution: an overview. Life 13:364. https://doi.org/10.3390/life13020364
Ben Maamar M, Beck D, Nilsson E et al (2022) Developmental alterations in DNA methylation during gametogenesis from primordial germ cells to sperm. iScience 25:103786. https://doi.org/10.1016/j.isci.2022.103786
Burruel V, Klooster KL, Chitwood J et al (2013) Oxidative damage to rhesus macaque spermatozoa results in mitotic arrest and transcript abundance changes in early embryos. Biol Reprod 89:72. https://doi.org/10.1095/biolreprod.113.110981
Cejko BI, Sarosiek B, Krejszeff S, Kowalski RK (2018) Multiple collections of common carp Cyprinus carpio L. semen during the reproductive period and its effects on sperm quality. Anim Reprod Sci 188:178–188. https://doi.org/10.1016/j.anireprosci.2017.12.002
Chao SB, Li JC, Jin XJ et al (2012) Epigenetic reprogramming of embryos derived from sperm frozen at -20°C. Sci China Life Sci 55:349–357. https://doi.org/10.1007/s11427-012-4309-8
Chatterjee A, Saha D, Niemann H et al (2017) Effects of cryopreservation on the epigenetic profile of cells. Cryobiology 74:1–7. https://doi.org/10.1016/j.cryobiol.2016.12.002
Chen M, Ruan R, Zhong X et al (2021) Comprehensive analysis of genome-wide DNA methylation and transcriptomics between ovary and testis in Monopterus albus. Aquac Res 52:5829–5839. https://doi.org/10.1111/are.15457
Cheng Y, Vechtova P, Fussy Z et al (2021) Changes in phenotypes and DNA methylation of in vitro aging sperm in common carp Cyprinus carpio. Int J Mol Sci 22:5925. https://doi.org/10.3390/ijms22115925
Cheng Y, Zhang S, Linhartová Z et al (2022a) Practical use of extender and activation solutions for short-term storage of common carp (Cyprinus carpio) milt in a hatchery. Aquac Rep 24:101160. https://doi.org/10.1016/j.aqrep.2022.101160
Cheng Y, Zhang S, Linhartová Z et al (2022b) Common carp (Cyprinus carpio) sperm reduction during short-term in vitro storage at 4 °C. Anim Reprod Sci 243:107017. https://doi.org/10.1016/j.anireprosci.2022.107017
Cheng Y, Waghmare SG, Zhang S et al (2023) Aging of common carp (Cyprinus carpio L.) sperm induced by short-term storage does not alter global DNA methylation and specific histone modifications in offspring. Aquaculture 571:739484. https://doi.org/10.1016/j.aquaculture.2023.739484
D’Occhio MJ, Campanile G, Zicarelli L et al (2020) Adhesion molecules in gamete transport, fertilization, early embryonic development, and implantation—role in establishing a pregnancy in cattle: a review. Mol Reprod Dev 87:206–222. https://doi.org/10.1002/mrd.23312
Darszon A, Labarca P, Nishigaki T, Espinosa F (1999) Ion channels in sperm physiology. Physiol Rev 79:481–510. https://doi.org/10.1152/physrev.1999.79.2.481
Daxinger L, Whitelaw E (2012) Understanding transgenerational epigenetic inheritance via the gametes in mammals. Nat Rev Genet 13:153–162. https://doi.org/10.1038/nrg3188
Deaton AM, Bird A (2011) CpG islands and the regulation of transcription. Genes Dev 25:1010–1022. https://doi.org/10.1101/gad.2037511
Depincé A, Gabory A, Dziewulska K et al (2020) DNA methylation stability in fish spermatozoa upon external constraint: Impact of fish hormonal stimulation and sperm cryopreservation. Mol Reprod Dev 87:124–134. https://doi.org/10.1002/mrd.23297
Dietrich MA, Judycka S, Słowińska M et al (2021) Short-term storage-induced changes in the proteome of carp (Cyprinus carpio L.) spermatozoa. Aquaculture 530:735784. https://doi.org/10.1016/j.aquaculture.2020.735784
Donkin I, Barrès R (2018) Sperm epigenetics and influence of environmental factors. Mol Metab 14:1–11. https://doi.org/10.1016/j.molmet.2018.02.006
Egger G, Liang G, Aparicio A, Jones PA (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429:457–463. https://doi.org/10.1038/nature02625
Fang X, Poulsen RR, Wang-Hu J et al (2016) Knockdown of DNA methyltransferase 3a alters gene expression and inhibits function of embryonic cardiomyocytes. FASEB J 30:3238–3255. https://doi.org/10.1096/fj.201600346R
Feng S, Jacobsen SE, Reik W (2010) Epigenetic reprogramming in plant and animal development. Science (80-) 330:622–627. https://doi.org/10.1126/science.1190614
Fitz-James MH, Cavalli G (2022) Molecular mechanisms of transgenerational epigenetic inheritance. Nat Rev Genet 23:325–341. https://doi.org/10.1038/s41576-021-00438-5
Gazo I, Shaliutina-Kolešová A, Dietrich MA et al (2015) The effect of reactive oxygen species on motility parameters, DNA integrity, tyrosine phosphorylation and phosphatase activity of common carp (Cyprinus carpio L.) spermatozoa. Mol Reprod Dev 82:48–57. https://doi.org/10.1002/mrd.22442
Gazo I, Naraine R, Lebeda I et al (2022) Transcriptome and proteome analyses reveal stage-specific DNA damage response in embryos of sturgeon (Acipenser ruthenus). Int J Mol Sci 23:6392. https://doi.org/10.3390/ijms23126392
Gerecke C, Schumacher F, Edlich A et al (2018) Vitamin C promotes decitabine or azacytidine induced DNA hydroxymethylation and subsequent reactivation of the epigenetically silenced tumour suppressor CDKN1A in colon cancer cells. Oncotarget 9:32822–32840. https://doi.org/10.18632/oncotarget.25999
Giassetti MI, Miao D, Law NC et al (2023) ARRDC5 expression is conserved in mammalian testes and required for normal sperm morphogenesis. Nat Commun 14:2111. https://doi.org/10.1038/s41467-023-37735-y
Gosálvez J, López-Fernández C, Hermoso A et al (2014) Sperm DNA fragmentation in zebrafish (Danio rerio) and its impact on fertility and embryo viability: implications for fisheries and aquaculture. Aquaculture 433:173–182. https://doi.org/10.1016/j.aquaculture.2014.05.036
He L, Liang X, Wang Q et al (2022) Genome-wide DNA methylation reveals potential epigenetic mechanism of age-dependent viral susceptibility in grass carp. Immun Ageing 19:28. https://doi.org/10.1186/s12979-022-00285-w
Herráez MP, Ausió J, Devaux A et al (2017) Paternal contribution to development: sperm genetic damage and repair in fish. Aquaculture 472:45–59. https://doi.org/10.1016/j.aquaculture.2016.03.007
Herranz-Jusdado JG, Gallego V, Rozenfeld C et al (2019) European eel sperm storage: optimization of short-term protocols and cryopreservation of large volumes. Aquaculture 506:42–50. https://doi.org/10.1016/j.aquaculture.2019.03.019
Jiang L, Zhang J, Wang JJ et al (2013) Sperm, but not oocyte, DNA methylome is inherited by zebrafish early embryos. Cell 153:773–784. https://doi.org/10.1016/j.cell.2013.04.041
Jones PA (2012) Functions of DNA methylation: Islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. https://doi.org/10.1038/nrg3230
Katayama S, Andou M (2021) Editing of DNA methylation using CRISPR/Cas9 and a ssDNA template in human cells. Biochem Biophys Res Commun 581:20–24. https://doi.org/10.1016/j.bbrc.2021.10.018
Kelley JL, Tobler M, Beck D et al (2021) Epigenetic inheritance of DNA methylation changes in fish living in hydrogen sulfide-rich springs. Proc Natl Acad Sci USA 118:e2014929118. https://doi.org/10.1073/pnas.2014929118
Khezri A, Narud B, Stenseth EB et al (2019) DNA methylation patterns vary in boar sperm cells with different levels of DNA fragmentation. BMC Genomics 20:897. https://doi.org/10.1186/s12864-019-6307-8
Krueger F, Andrews SR (2011) Bismark: a flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27:1571–1572. https://doi.org/10.1093/bioinformatics/btr167
Kurdyukov S, Bullock M (2016) DNA methylation analysis: choosing the right method. Biology (basel) 5:3. https://doi.org/10.3390/biology5010003
Labbé C, Robles V, Herraez MP (2017) Epigenetics in fish gametes and early embryo. Aquaculture 472:93–106. https://doi.org/10.1016/j.aquaculture.2016.07.026
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. https://doi.org/10.1038/nmeth.1923
Lee TF, Zhai J, Meyers BC (2010) Conservation and divergence in eukaryotic DNA methylation. Proc Natl Acad Sci USA 107:9027–9028. https://doi.org/10.1073/pnas.1005440107
Linhart O, Rodina M, Gela D et al (2005) Spermatozoal competition in common carp (Cyprinus carpio): What is the primary determinant of competition success? Reproduction 130:705–711. https://doi.org/10.1530/rep.1.00541
Linhart O, Rodina M, Kašpar V (2015) Common carp (Cyprinus carpio Linneaus, 1758) male fertilization potency with secure number of spermatozoa per ova. J Appl Ichthyol 31:169–173. https://doi.org/10.1111/jai.12736
Liu Z, Zhou T, Gao D (2022) Genetic and epigenetic regulation of growth, reproduction, disease resistance and stress responses in aquaculture. Front Genet 13:994471. https://doi.org/10.3389/fgene.2022.994471
Mao X, Cai T, Olyarchuk JG, Wei L (2005) Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21:3787–3793. https://doi.org/10.1093/bioinformatics/bti430
Metzger DCH, Schulte PM (2016) Epigenomics in marine fishes. Mar Genomics 30:43–54. https://doi.org/10.1016/j.margen.2016.01.004
Murphy PJ, Wu SF, James CR et al (2018) Placeholder nucleosomes underlie germline-to-embryo DNA methylation reprogramming. Cell 172:993-1006.e13. https://doi.org/10.1016/j.cell.2018.01.022
Nakamura M, Gao Y, Dominguez AA, Qi LS (2021) CRISPR technologies for precise epigenome editing. Nat Cell Biol 23:11–22. https://doi.org/10.1038/s41556-020-00620-7
Natarajan VP, Zhang X, Morono Y et al (2016) A modified SDS-based DNA extraction method for high quality environmental DNA from seafloor environments. Front Microbiol 7:986. https://doi.org/10.3389/fmicb.2016.00986
Niu J, Wang X, Liu P et al (2022) Effects of cryopreservation on sperm with cryodiluent in viviparous black rockfish (Sebastes schlegelii). Int J Mol Sci 23:3392. https://doi.org/10.3390/ijms23063392
Ohka F, Natsume A, Motomura K et al (2011) The global DNA methylation surrogate LINE-1 methylation is correlated with MGMT promoter methylation and is a better prognostic factor for glioma. PLoS ONE 6:e23332. https://doi.org/10.1371/journal.pone.0023332
Ortiz-Rodriguez JM, Ortega-Ferrusola C, Gil MC et al (2018) Transcriptome analysis reveals that fertilization with cryopreserved sperm downregulates genes relevant for early embryo development in the horse. PLoS ONE 14:e0213420. https://doi.org/10.1371/journal.pone.0213420
Pan Y, Chen L, Cheng J et al (2021) Genome-wide DNA methylation profiles provide insight into epigenetic regulation of red and white muscle development in Chinese perch Siniperca chuatsi. Comp Biochem Physiol Part - B Biochem Mol Biol 256:110647. https://doi.org/10.1016/j.cbpb.2021.110647
Park Y, Wu H (2016) Differential methylation analysis for BS-seq data under general experimental design. Bioinformatics 32:1446–1453. https://doi.org/10.1093/bioinformatics/btw026
Perchec G, Cosson MP, Cosson J et al (1996) Morphological and kinetic changes of carp (Cyprinus carpio) spermatozoa after initiation of motility in distilled water. Cell Motil Cytoskeleton 35:113–120. https://doi.org/10.1002/(sici)1097-0169(1996)35:2%3c113::aid-cm4%3e3.3.co;2-l
Pérez-Cerezales S, Martínez-Páramo S, Cabrita E et al (2009) Evaluation of oxidative DNA damage promoted by storage in sperm from sex-reversed rainbow trout. Theriogenology 71:605–613. https://doi.org/10.1016/j.theriogenology.2008.09.057
Pérez-Cerezales S, Martínez-Páramo S, Beirão J, Herráez MP (2010) Fertilization capacity with rainbow trout DNA-damaged sperm and embryo developmental success. Reproduction 139:989–997. https://doi.org/10.1530/REP-10-0037
Potok ME, Nix DA, Parnell TJ, Cairns BR (2013) Reprogramming the maternal zebrafish genome after fertilization to match the paternal methylation pattern. Cell 153:759–772. https://doi.org/10.1016/j.cell.2013.04.030
Schumacher F, Herrmann K, Florian S et al (2013) Optimized enzymatic hydrolysis of DNA for LC-MS/MS analyses of adducts of 1-methoxy-3-indolylmethyl glucosinolate and methyleugenol. Anal Biochem 434:4–11. https://doi.org/10.1016/j.ab.2012.11.001
Shaliutina A, Hulak M, Gazo I et al (2013) Effect of short-term storage on quality parameters, DNA integrity, and oxidative stress in Russian (Acipenser gueldenstaedtii) and Siberian (Acipenser baerii) sturgeon sperm. Anim Reprod Sci 139:127–135. https://doi.org/10.1016/j.anireprosci.2013.03.006
Shazada NE, Alavi SMH, Siddique MAM et al (2024) Short-term storage of sperm in common carp from laboratory research to commercial production: a review. Rev Aquac 16:174–189. https://doi.org/10.1111/raq.12827
Strömqvist M, Tooke N, Brunström B (2010) DNA methylation levels in the 5’ flanking region of the vitellogenin I gene in liver and brain of adult zebrafish (Danio rerio)-Sex and tissue differences and effects of 17α-ethinylestradiol exposure. Aquat Toxicol 98:275–281. https://doi.org/10.1016/j.aquatox.2010.02.023
Vehniäinen ER, Bremer K, Scott JA et al (2016) Retene causes multifunctional transcriptomic changes in the heart of rainbow trout (Oncorhynchus mykiss) embryos. Environ Toxicol Pharmacol 41:95–102. https://doi.org/10.1016/j.etap.2015.11.015
Wang X, Bhandari RK (2019) DNA methylation dynamics during epigenetic reprogramming of medaka embryo. Epigenetics 14:611–622. https://doi.org/10.1080/15592294.2019.1605816
Woolner S, Martin P (2006) Embryo morphogenesis and the role of the actin cytoskeleton. Adv Mol Cell Biol 37:251–283. https://doi.org/10.1016/S1569-2558(06)37012-9
Wyck S, Herrera C, Requena CE et al (2018) Oxidative stress in sperm affects the epigenetic reprogramming in early embryonic development. Epigenetics Chromatin 11:60. https://doi.org/10.1186/s13072-018-0224-y
Yang Y, Wang T, Yang S et al (2021) The effects of storage in vitro on functions, transcriptome, proteome, and oxidation resistance of giant grouper sperm. Front Mar Sci 8:716047. https://doi.org/10.3389/fmars.2021.716047
Young MD, Wakefield MJ, Smyth GK, Oshlack A (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 11:R14. https://doi.org/10.1186/gb-2010-11-2-r14
Zhang S, Cheng Y, Věchtová P et al (2023a) Potential implications of sperm DNA methylation functional properties in aquaculture management. Rev Aquac 15:536–556. https://doi.org/10.1111/raq.12735
Zhang S, Cheng Y, Alavi SMH et al (2023b) Elevated temperature promotes spermatozoa motility kinetics and fertilizing ability following short-term storage: an implication for artificial reproduction of common carp Cyprinus carpio in a hatchery. Aquaculture 565:739126. https://doi.org/10.1016/j.aquaculture.2022.739126
Zhou H, Zhuang ZX, Sun YQ et al (2019) Changes in DNA methylation during epigenetic-associated sex reversal under low temperature in Takifugu rubripes. PLoS ONE 14:e0221641. https://doi.org/10.1371/journal.pone.0221641
Acknowledgements
The authors thank Susanne Scheu (Freie Universität Berlin) for her excellent technical assistance with the DNA work-up for LC-MS/MS measurements.
Funding
This study was funded by the Ministry of Education, Youth and Sports of the Czech Republic (LRI CENAKVA, LM2023038), by project Biodiversity (CZ.02.1.01./0.0/0.0/16_025/0007370), by the Grant Agency of the University of South Bohemia in České Budějovice (107/2022/Z), by the Czech Science Foundation (23-06426S) and by the National Agency for Agriculture Research, Czech Republic (QK21010141).
Author information
Authors and Affiliations
Contributions
YC and OL conceived and designed the experiments. OL supervised the study and provided the funding. YC, SZ, PV, FS, IG, BK, AD, VR, MR and ZL carried out the experiments and collected data. YC and RN performed statistical analyses and generated figures and tables. YC, RN, SGW, SMHA and OL wrote the manuscript. YC, SZ, RN, PV, PL, JS, SMHA, CL and OL revised and reviewed the manuscript. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cheng, Y., Zhang, S., Nayak, R. et al. Fertilization by short-term stored sperm alters DNA methylation patterns at single-base resolution in common carp (Cyprinus carpio) embryos. Rev Fish Biol Fisheries (2024). https://doi.org/10.1007/s11160-024-09866-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11160-024-09866-y