Abstract
Clean and sustainable H2 production is crucial to a carbon–neutral world. H2 generation by Chlamydomonas reinhardtii is an attractive approach for solar-H2 from H2O. However, it is currently not large-scalable because of lacking desirable strains with both optimal H2 productivity and sufficient knowledge of underlying molecular mechanism. We hereby carried out extensive and in-depth investigations of H2 photoproduction of hpm91 mutant lacking PGR5 (Proton Gradient Regulation 5) toward its up-scaling and fundamental mechanism issues. We show that hpm91 is at least 100-fold scalable (up to 10 L) with continuous H2 collection of 7287 ml H2/10L-HPBR in averagely 26 days under sulfur deprivation. Also, we show that hpm91 is robust and active during sustained H2 photoproduction, most likely due to decreased intracellular ROS relative to wild type. Moreover, we obtained quantitative proteomic profiles of wild type and hpm91 at four representing time points of H2 evolution, leading to 2229 and 1350 differentially expressed proteins, respectively. Compared to wild type, major proteome alterations of hpm91 include not only core subunits of photosystems and those related to anti-oxidative responses but also essential proteins in photosynthetic antenna, C/N metabolic balance, and sulfur assimilation toward both cysteine biosynthesis and sulfation of metabolites during sulfur-deprived H2 production. These results reveal not only new insights of cellular and molecular basis of enhanced H2 production in hpm91 but also provide additional candidate gene targets and modules for further genetic modifications and/or in artificial photosynthesis mimics toward basic and applied research aiming at advancing solar-H2 technology.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hydrogen (H2) derived from H2O is a clean and versatile energy carrier that could be obtained sustainably through sunlight-driven chemical and biological means such as photocatalysis and algal photoproduction (Bayro-Kaiser and Nelson 2017; Nishiyama et al. 2021). Under anaerobic condition, H2 photoproduction occurs in microalgae such as Chlamydomonas reinhardtii (henceforth referred to as Chlamydomonas) via two distinct light-driving pathways dependent on the activities of photosystems and (Fe–Fe) hydrogenases (Chochois et al. 2009; Fouchard et al. 2005). H2 formation is through the catalytic activity of (Fe–Fe) hydrogenases (Forestier et al. 2003) induced under anoxia, using mainly photosynthetic electrons on the acceptor side of photosystem I (PSI) in reduction of protons into H2. As a result, renewable H2 production is achieved in the organisms using solar energy and electrons derived from photosynthetic water-splitting reaction. However, H2 evolution by wild-type Chlamydomonas strains is only transient with small amounts, probably due to high sensitivity of the hydrogenases to O2 released from PSII (Ghirardi 2015). Significant progress was made mainly by Melis et al. (Melis et al. 2000) and Kruse et al. (Kruse et al. 2005) with development of the sulfur-deprivation method for sustained H2 evolution and establishment of stm6 mutant that produces 540 ml of H2 /1L culture up to 14 days under such condition, respectively.
Making use of genomic sequence information (Merchant et al. 2007) and various tools of systems biology (Nguyen et al. 2008; Matthew et al. 2009; Chen et al. 2010; Toepel et al. 2013, numerous Chlamydomonas mutants with increased H2 production have been obtained. Several of them are reasonably well studied such as the truncated light-harvesting antenna mutant (tla1), D1 mutant (Kosourov et al. 2011; Scoma et al. 2012), and the pgr mutants named as pgrl1 (proton gradient regulation like 1), pgr5 (proton gradient regulation 5), and hpm91 (Tolleter et al. 2011; Steinbeck et al. 2015; Chen et al. 2016; 2019). The pgr mutants appeared highly favorable because their target genes (Pgrl1, Pgr5) are involved in the PGR5-dependent path of photosynthetic cyclic electron flow (CEF), which is the major branch of CEF (Takahashi et al. 2013; Schwenkert et al. 2022). Inverse relationship of CEF and H2 evolution was firstly revealed in the study of stm6 mutant lacking MOC1 (an assembly factor of the mitochondrial respiratory chain) (Schonfeld et al. 2004; Kruse et al. 2005) as well as the observation of increased H2 production upon addition of the CEF inhibitor antimycin A (Antal et al. 2009). More experimental evidence was also obtained via analysis of pgr (pgrl1, pgr5, hpm91) as well as fnr (ferredoxin-NADPH reductase) mutants (Yacoby et al. 2011; Sun et al. 2013). PGRL1 and FNR have been identified in the CEF supercomplex (Iwai et al. 2010) and PGR5 is known being an important protein involved in the CEF branch (Munekage et al. 2002; Suorsa et al. 2012), for which the mechanistic mode is currently not completely understood (Schwenkert et al. 2022).
Remarkably, the hpm91 mutant sustains H2 production for 25 days with 30-fold yield increase relative to wild type (Chen et al. 2016). Although a negative correlation was already revealed between PGR5 levels and H2 production, and the prolonged H2 evolution was mainly attributed to the enhanced anti-ROS capability protecting the photosynthetic electron transport chain from photooxidative damage, questions arise as (i) whether the phenotype of hpm91 is stable in large scale setting-ups? (ii) how intracellular ROS content of hpm91 is altered during sulfur-deprived H2 photoproduction? (iii) what are the proteomic characteristics of hpm91 under such conditions; (iv) is it possible to obtain high H2-producing mutants excess to hpm91? We report here the performance of hpm91 in a large scale (up to 10 L) H2-photobioreactor (HPBR) systems. We also describe insightful findings of hpm91 based on quantitative proteomic analysis. We highlight the hpm91 as a valuable algal strain not only for basic research of understanding the molecular mechanisms of bio-H2 photoproduction but also for development of economically viable solar-powered H2 production systems.
Materials and methods
Algal cultivation and H2 photoproduction
Wild-type Chlamydomonas strain CC400 was purchased from the Chlamydomonas Center (www.Chlamy.org) and the mutant strain hpm91 was isolated in our laboratory and previously reported (Chen et al. 2016; 2019). Genetic analysis and phenotypic rescue of several fully complemented strains as well as immunoblot detection suggest that the loss of Pgr5 gene is responsible for the H2-overproducing phenotype of hpm91 (Chen et al. 2016).
H2 production was induced using sulfur-deprivation method (Melis et al. 2000) with minor modifications (Chen et al. 2010; Sun et al. 2013). Algal cells were grown in TAP medium (Gorman and Levine 1965) at 25 °C under continuous light (60 µmol photons m−2 s−1) until mid-exponential phase. Cells were pelleted and washed once with sulfur-depleted TAP medium followed by resuspending in the medium with desired cell density of 20- (for 10L-HPBR) and 25 μg ml−1 chlorophyll (a and b) (Arnon 1949), respectively. H2 evolution was achieved via transferring the culture into small H2 photobioreactor (100 ml, light path 4.5 cm) for biochemical analysis and large gas-tight glass bottles (large HPBR, upto 10 L; light path 10 cm for 1-3L and 22 cm for 10L, respectively) connected by a Teflon tube to storage glass cylinder for scaling-up studies followed by magnetic-bar-stirring cultivation at the same condition mentioned above (for small HPBR) or upto 230 µmol photons m−2 s−1 (for large HPBR), respectively. H2 gas accumulated in the headspace of the small HPBR was measured by gas chromatograph (GC-2014, Shimadzu, Japan) in the way as previously described (Sun et al. 2013; Zhao et al. 2013; Chen et al. 2016; 2019). The evolved H2 gas from the headspace of large HPBR was collected in inverted graduated cylinders and measured by the displacement of water (Melis et al. 2000).
Intracellular ROS and cell growth analysis
Intracellular ROS content was determined using a MoFlo XDP high-speed flow cytometer (Beckman-Coulter, Inc. USA) by following the manufacturer’s instructions. Samples were prepared in an anoxia workstation (Longyao, LAI-3 T; Shanghai, China) as (Chen et al. 2019). Cells were incubated with 10 mM H2DCFDA at 25 °C for 30 min in dark then examined by the flow cytometer. For detection of DCF green fluorescence, wavelength of excitation at 488 nm and emission at 510 to 550 nm was used. Rosup provided by a ROS assay kit (Beyotime Institute of Biotechnology, Haimen, China) was used as a positive control. Average fluorescence intensity of DCF from 3 × 105 cells was recorded and data acquisition and analysis were carried out using Summit 5.2 software (Beckman-Coulter, Inc. USA). Morphology of algal cells was examined and photographed with a differential interference contrast microscopy (DIC) (Leica DM4500, Germany). Culture density were determined by cell counting using a hemocytometer.
iTRAQ proteomics and data analysis
Protein extraction and sample preparation for iTRAQ labeling was done as described (Chen et al. 2010) with minor modifications (Ge et al. 2017). Frozen cells suspended in ice-cold extraction buffer were disrupted with glass beads (diameter 150–212 μm, Sigma) via vortexing 30 s/5 cycles/each with 1-min break on ice. After removed unbroken cells and insoluble debris, total proteins in the supernatant were precipitated with ice-cold 10% trichloroacetic acid (TCA) in acetone at − 20 °C. Protein pellets were washed with acetone by centrifugation and vacuum-dried. The proteins were resolubilized with 4% sodium dodecyl sulfate (SDS) in 0.1 M Tris–HCl, pH 7.6. Protein concentration was determined using BCA protein assay kit (Thermo Scientific, Rockford, IL).
Trypsin digestion of proteins was performed using the filter-aided sample preparation (FASP) method with slight modifications (Wisniewski et al. 2009). The resulting tryptic peptides were labeled with 8-plex iTRAQ reagents (AB Sciex Inc., MA, USA) alternatively by following manufacturer's manual. The iTRAQ labeled samples were mixed together with equal ratios in amount, and the mixture was concentrated with a SpeedVac in subjection for fractionation by HPLC (Waters, e2695 separations) coupled with a phenomenex gemini-NX 5u C18 column (250 × 3.0 mm, 110 Å) (Torrance, CA, USA). The samples were then separated using a 97 min basic RP-LC gradient as described (Udeshi et al. 2013) and a flow rate of 0.4 mL/min was used. The separated samples were collected into 10 fractions and vacuum-dried prior to LC–MS/MS analysis by a TripleTOF 5600 mass spectrometer (AB SCIEX) coupled online to an Eksigent nanoLC Ultra in Information Dependent Mode. Peptides were separated on a C18 column (Acclaim PepMap C18, 250 mm × 75 μm × 5 μm, 100 Å, Dionex) with a 90 min nonlinear gradient of buffer B (100% ACN, 0.1% FA) from 3 to 30%. The gradient was set as 3–8% B for 10 min, 8–20% B for 60 min, 20–30% B for 8 min, 30–100% B for 2 min, and 100% B for 10 min at a flow rate of 300 nl/min. MS spectra survey scan was done across mass range of 350 to 1500 m/z and the spectra data were acquired at resolution 30,000 with 250 ms accumulation per spectrum. 25 most intense ions from each MS scan were chosen for fragmentation from each MS spectrum with 2 s minimum accumulation time for each precursor and dynamic exclusion for 18 s. Tandem mass spectra were recorded in high sensitivity mode (resolution > 15,000) with rolling collision energy on and iTRAQ reagent collision energy adjustment on.
Peptide and protein identification and quantification was performed with the ProteinPilot 4.5 software (AB SCIEX) using the Paragon database search algorithm. The Chlamydomonas proteome sequences downloaded from UniProt (dated 2–07-2016) were used for the database searching with manual editing using the information (updated on 11–12-2019) in Uniprot database. The false discovery rate (FDR) analysis was performed using the software PSPEP integrated with the ProteinPilot. Confidence of quantitation for differentially expressed proteins was analyzed with the ProteinPilot Descriptive Statistics Template (PDST) (beta v3.07p, AB SCIEX). Gene ontology (GO) enrichment analysis of proteins was performed using DAVID Bioinformatic Resources 6.8 (https://david.ncifcrf.gov/summary.jsp) with the following parameters: ‘Gene List Enrichment,’ ‘Chlamydomonas reinhardtii species,’ ‘Uniprot Accession.’ Functional annotation and classification of the differentially expressed proteins was mainly based on the search results of ‘Biological processes’ in DAVID analysis followed by manual editing using the updated Chlamydomonas genome data in NCBI (updated on 30–01-2018). Differentially expressed proteins were selected with p value < 0.05 and cut-off > 1.2 and < 0.83 as up-and down-regulated, respectively.
Generation and screening of hpm91-derived mutants
Insertion mutant library was constructed by transformation of hpm91 using the glass bead method with KpnI-linearized plasmid pDble containing the ble gene conferring zeocin resistance (Kindle 1990). Transformants growing on TAP plates with 10 μg mL−1 zeocin (Solarbio) were isolated. After cultured on TAP-S plates for one week, mutants were screened based on increased Y(II) values relative to hpm91 using Maxi-Imaging PAM chlorophyll fluorometer (Walz, Germany) as previously mentioned (Zhao et al. 2017) followed by H2 measurements by GC analysis (Sun et al. 2013; Chen et al. 2019).
Results and discussion
H2 production of hpm91 mutant was large-scalable
Based on experimental results reported so far, hpm91 appears remarkable among the single mutants deficient in pgr genes of Chlamydomonas with enhanced H2 photoproduction under sulfur-deprived condition (Tolleter et al. 2011; Steinbeck et al. 2015; Chen et al. 2016; Ho et al. 2022). To determine its potential toward application, we carried out scaling-up and gas-handling experiments in the laboratory using hpm91 (Fig. 1; Movie S1). Initially, the experiments were performed with H2-photobioreactors (HPBR) of scaling-up 10-, 20-, and 30-times (from 100 ml to 1L, 2L, and 3L) with chlorophyll concentration of 25 µg/ml as previously used (Chen et al. 2016; Fig. 1a). Although the yield of H2 production in hpm91 was slightly lower or comparable to the original pgr5 mutant in 1-L set-ups (Steinbeck et al. 2015; Ho et al. 2022), we found the highest yield of H2 from 3L-HPBR and a positive correlation between the yield and size of HPBR (Fig. 1b). These results encouraged us to extend the scaling-up directly to 10L-HPBR (headspace 1700 ml, light path 22 cm, illuminated on 3 sides) and to investigate its H2 production profiles. To reduce potential shading effects (Kosourov et al. 2002; Hemschemeier et al. 2009) and low-light stress on algal cells, we reasonably reduced initial chlorophyll concentration (ICC) to 20 µg/ml of cell suspension and cultured under increased light irradiance (130 µmol photons m−2 s−1) for H2 production.
H2 production of hpm91 in 1 to 10 L-HPBR under sulfur deprivation a Photograph of 1 to 3 L-HPBR (optical path 10 cm, 100 µmol photons m−2 s−1.) taken at 10 day of sulfur deprivation. b Correlation between H2 output of hpm91 and size of HPBR. Standard deviations were estimated from three biological replicates. c Photograph of wild type and hpm91 in 10L-HPBR taken at 4 day of sulfur deprivation. d Comparison of H2 output profiles of hpm91 in 10L-HPBR under light intensity of 130 (in black) and 230 µmol photons m−2 s−1 (in red). The data are from three independent experiments
Figure 1c compares H2 photoproduction of hpm91 and wild type in 10L-HPBR under identical cultural condition. In contrast to wild type, which generated few gas bubbles with collectable H2 quantity negligible, we observed a bulk of H2 bubbles in hpm91 after 3–4 days of sulfur deprivation (Fig. 1c) with the gas collectable for up to 33 days (Table S1). To confirm these results, experiments were repeated using different batch of algal cells. The data shows that continuous H2 collection from hpm91 could be achieved at an average of 28 (28 ± 4) days under light irradiance of 130 µmol photons m−2 s−1 (Table S1), which was the longest duration of H2 production under sulfur deprivation reported thus far for Chlamydomonas. These results promoted us to explore more possibilities to enhance H2 production of hpm91 in 10L-HPBR.
Indeed, further increase of light intensity to 230 µmol photons m−2 s−1 apparently enhanced H2-producing capability of hpm91 (Table S1). Our data shows that while the durations of H2 production under both light intensities were roughly comparable, the average yield of H2 obtained under the elevated light irradiance (230 µmol photons m−2 s−1) was 7287 (7287 ± 986) ml/10L-HPBR, which was 50.7% higher than that under lower light condition (Table S1). To obtain more information of H2 photoproduction, we compared time course of H2 collection in hpm91 grown under such conditions (Fig. 1d). Considering that in both cases H2 output was relatively stable around 25 day of sulfur deprivation, H2 evolution profiles of hpm91 during this period were compared. As shown in Fig. 1d, the general kinetic pattern was largely similar but significantly higher average rate of H2 production was found (710 ml d−1 vs 420 ml d−1) for those cultured under increased light. Because no further scaling-up data is available thus far for the original pgr5 mutant, our present results of hpm91 obtained in the large scale (2–10 L) are novel among the pgr5 mutants. Further experiments verified its essentially pure H2 output which enabled us to make a H2 fuel cell-powered toycar drive using ambient air directly (Movie S1). These results allowed us to address that hpm91 is a potent H2 producer for further genetic engineering toward large-scale H2 photoproduction.
Decreased intracellular ROS in hpm91 during sustained H2 production
Based on visual comparison of cell suspension, cell growth of hpm91 was always better than wild type in both small (100 ml) and large HPBR (1 to 10 L) especially during prolonged period of H2 production. To understand the basis of cell biology, we investigated changes of cell morphology and viability in hpm91 committed to H2 production. Since the small HPBR system was experimentally more efficient, the system was of choice for comparative studies between wild type and hpm91 during 120 h of H2 production (Fig. 2).
Comparison of intracellular ROS contents of hpm91 and wild type during 120 h of sulfur-deprived H2 production process a and b Cell morphology and proliferation of the two strains. c and d Scatter diagram and quantification of intracellular ROS contents in the two strains. Sample preparation was performed under anoxia and ROS levels were determined using a MoFlo XDP high-speed flow cytometer (Beckman-Coulter, Inc. USA) by following the manufacturer’s instructions. Average fluorescence intensity of DCF from 3 × 105 cells was recorded and data acquisition and analysis were carried out using Summit 5.2 software. Experiments were repeated three times with similar results. * and ** refer to p-values < 0.05 and < 0.01 in Student’s t-test, respectively
Figure 2a shows that wild-type cells became largely translucent, whereas no such changes were observed in hpm91. This remarkable morphological alteration of wild-type cells was highly similar to earlier observations which was mainly attributed to the substantial loss of endogenous starch and a declined cell viability under such condition (Zhang et al. 2002). Also, culture density of hpm91 was more stable after 24 h and remained significantly higher than wild type at the end of the measurements (120 h) (Fig. 2b). Moreover, we found decreased portion of dead cells in the culture of hpm91 relative to wild type (Fig. S1). These results are clear indications of robustness of hpm91 cells during sulfur-deprived H2 production.
To further elucidate this, we measured intracellular ROS contents of hpm91 and wild type under such conditions (Fig. 2c and d). ROS levels were determined using a MoFlo XDP high-speed flow cytometer (Beckman-Coulter, Inc. USA) by following the manufacturer’s instructions. Sample preparation was performed under anoxia as described (Chen et al. 2019). Average fluorescence intensity of DCF from 3 × 105 cells was recorded and data acquisition/analysis were carried out using Summit 5.2 software (Beckman-Coulter, Inc. USA) (Fig. 2c). Our data shows that intracellular ROS in hpm91 remained significantly lower than wild type during H2 production. At 120 h, the amount of ROS in hpm91 was only 1/3 of that in wild type (Fig. 2d). Earlier physiological and biochemical studies strongly implicate occurrence of oxidative stress in Chlamydomonas wild-type and hpm91-mutant cells during sulfur-deprived H2 photoproduction (Sáenz et al. 2015; Chen et al. 2016; Kosourov et al. 2017). In this work, we demonstrated that cell viability was negatively correlated with intracellular ROS content (Fig. 2; Fig. S1), providing in vivo evidence of toxic effect of excess ROS on cell biology.
Overview of proteome changes in wild type and hpm91 under sulfur deprivation
To understand the molecular mechanism behind the remarkable phenotype of hpm91, we carried out iTRAQ proteomics of hpm91 and wild-type cells during 120 h of H2 production (Fig. 3). As presented in Fig. 3a, samples were taken at four different time points (0, 24, 72 and 120 h) and total proteins were extracted for protein identification and quantification. A total of 3798 proteins with quantitative data were confidently identified (FDR < 1%) as listed in (Dataset 1). Identification was mostly based on minimum of two peptide-hits per protein but considering several proteins such as hydrogenase3 (Hyd3) were probably involved H2 metabolism and numerous functionally important small-sized and/or membrane proteins may possess only one identifiable tryptic peptide in MS analysis, those identified with one-peptide hit were also reasonably included (Dataset 1).
Overview of iTRAQ proteomics of hpm91 and wild type during 120 h of sulfur-deprived H2 production a Schematic presentation of iTRAQ experimental design. b Hierarchy clustering analysis showing high reproducibility of protein quantitation. c Number of differential expressed proteins in hpm91 and wild type using a cutoff of 1.2-fold change with significance (p < 0.05). d KEGG analysis of group I proteins in (c) showing major proteome changes caused by PGR5 deletion in Chlamydomonas. DAVID Bioinformatic Resources 6.8 (https://david.ncifcrf.gov/summary.jsp) was used
Reproducibility of protein quantitation was verified by hierarchical clustering analysis (Perseus _1.6.0.7) of these proteins obtained with three biological replicates (Fig. 3b). To confirm functional impairment of hpm91 in cyclic electron transfer (CEF), we compared the CEF rates in both strains. The data showed that CEF of hpm91 was significantly decreased relative to wild type (Fig. S2a). To be more certain with the genetic background of the strains, we performed high-throughput genomic sequencing for wild type (CC400, 137c) and the pgr5 mutants (hpm91, pgr5). The data was deposited into CNGB Sequencing Archive1of CNGBdb2 database (accession No. CNP0002674). Further analysis of reads coverage validates previous mutation mapping (Johnson et al. 2014; Chen et al. 2016) and showed large deletions in PGR5-containing region of hpm91 and pgr5 mutants (Supplemental Fig. S2b). Together with the immuno-blot results showing no detectable PGR5 but presence of PGRL1 and FNR in hpm91 (Chen et al. 2016), we clarify that the impaired CEF of hpm91 is attributed to loss of PGR5. Differentially expressed proteins were determined using a cutoff of 1.2-fold change with significance (p < 0.05), leading to three groups consisting of 529 (group I), 2229 (group II), and 1350 (group III) proteins (Fig. 3c) listed in (Datasets 2–4). These correspond to three comparisons, i.e., hpm91 at 0 h vs wild-type at 0 h, any time of- wild type vs wild-type at 0 h, and any time of-hpm91 vs hpm91 at 0 h, representing differentially expressed proteins caused by deletion of PGR5 and by sulfur-deprived anoxia in wild type and hpm91, respectively.
To understand functional significance of the differentially expressed proteins in each group, KEGG and gene ontology (GO) analysis was performed using DAVID Bioinformatic Resources 6.8 (https://david.ncifcrf.gov/summary.jsp), yielding 4 enriched KEGG pathways for group I (Fig. 3d), 30 and 29 enriched biological process (GOPB) for the later two groups, respectively (Fig. 4a, Datasets 5 and 6). Because numerous proteins in the latter two enrichments were multiply or/and with error assignments, we reasonably delineated them into 8 and 9 major groups as (Wang et al. 2012) and shown in (Tables S2 and S3), respectively.
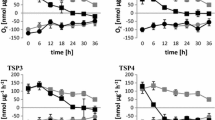
Proteomic characteristics of hpm91 during sustained H2 production a Comparison of GOBP enrichments in hpm91 and wild type during 120 h of sulfur deprivation. b Major proteome feature of hpm91 under sulfur-deprived condition. Volcano plot shows major changes in carbon metabolism of hpm91 during 120 h of sulfur deprivation (upleft panel). Comparison of average fold-change of photosynthetic proteins in hpm91 and wild type at 120 h sulfur deprivation (upright panel). Dynamic changes of redox proteins in hpm91 during 120 of sulfur deprivation (downleft panel). Schematic illustration of N- and S-metabolic features in hpm91 under sulfur-deprived condition (downright panel)
It can be seen in Table 1, deletion of PGR5 caused significant changes in four pathways in Chlamydomonas. Compared to wild type, all the ribosomal proteins and most of those related to nitrogen metabolism were higher-expressed in hpm91. The latter may implicate enhanced nitrogen metabolism in hpm91. To test this possibility, we compared phenotype of the two strains under N-starved stress condition. The experimental data showed that both cell growth and photosynthetic capability of hpm91 was indeed better than wild type (Fig. S2c), revealing another impact of PGR5 on chloroplast biology. Apparently, further investigations are required to uncover the molecular mechanism behind this phenotype. More interestingly, we found all the photosynthetic antenna proteins (except for LHCA3) were lower-expressed in hpm91. Because those account for more than 50% of LHCI and LHCII proteins in Chlamydomonas (Shen et al. 2019; Su et al. 2019; Suga et al. 2019), our finding of their reduced levels could be an indication of a smaller photosynthetic antenna in hpm91 than wild type. Notably, among the proteins involved in sulfur metabolism only APS reductase APR1 encoded by APR1/MET16 (Gutierrez-Marcos et al. 1996; Setya et al. 1996), known to be involved in sulfur-starvation response (Ravina et al. 2002; Zhang et al. 2004) was higher-expressed in hpm91 (Table 1).
Comparison of the results in Tables S2 and S3 revealed similarities in 5 functional groups, i.e., translation, protein folding, intracelluar protein trafficking, response to cytokinin, and ATP hydrolysis/production in the two strains. However, differences were also revealed in 4 of those corresponding to carbon metabolism, photosynthetic antenna, cell redox homeostasis/anti-oxidative systems, and nitrogen- and sulfur metabolisms. These are the major proteomic characteristics of hpm91-cells committed to sulfur-deprived H2 production, which are described/discussed in the following section.
Proteomic characteristics of hpm91 during sustained H2 production
Loss of PGR5 in hpm91 causes compromised primary carbon metabolism
As can be seen in Fig. 4a, six biological processes were only enriched in hpm91, i.e., ‘carbohydrate metabolism,’ ‘gluconeogenesis,’ ‘fructose 6-phosphate metabolic process,’ ‘glyoxylate cycle,’ ‘cellular amino acid metabolic process,’ and ‘terpenoid biosynthetic process,’ with the first one appeared within top-10 rankings. This implicates that loss of PGR5 caused more profound alterations of primary carbon metabolism in hpm91 than wild type during H2 production. Considering metabolic relevance in Chlamydomonas under anoxia (Yang et al. 2015), the group ‘tricarboxylic acid cycle’ was combined with the first four groups in (Table S3). While many of them overlapped with wild type, 16 proteins were exclusively revealed in hpm91 during sustained H2 production. These include the key proteins in the regeneration pathway of CBB cycle (SEBP1), glyoxylate cycle (ICL1), gluconeogenesis (PCK1b |PCK1a), oxidative PPP pathway (TAL1, TAL2), fermentative pathways (PFK1, PFK2, PGI1, PFL1), starch metabolism (AMYA2, AMY-like protein, GHL1, ATF1, PHOB, GPM2), and TCA cycle (IDH1), showing decreased and increased trends for the most proteins in the first four and the latter three pathways, respectively (Fig. 4b, upleft pannel). It was earlier reported that loss of PFL1 decreased H2 photoproduction (Philipps et al. 2011). Our finding of increased amount of PFL1 in hpm91 was in line with this and may suggest a partial contribution of PFL1 to its enhanced H2 production. Also, we found two alpha-amylases, glucosamine-fructose-6-phosphate aminotransferase (ATF1) and phosphorylase PHOB increased at an average of 1.86 to 5.02-fold in hpm91 during H2 production (Table S3). These enzymes are known to be essential for starch metabolism (Weigelt et al. 2009). While accumulation of PHOB could be correlated to the marked increase of starch contents in hpm91 (Chen et al. 2016), elevated level of the amylases was somehow intriguing because starch breakdown in hpm91 was found significantly less than wild type (Chen et al. 2016), excluding the major contribution of ‘indirect pathway’ on the prolonged H2 production phenotype of hpm91. Moreover, our data revealed down-regulated key enzymes in or related to CBB cycle (PRK1, FBP1, SEBP1) in hpm91, suggesting that the main route of CO2 fixation was largely repressed toward H2 production. It is possible that the increased levels of amylases as well as GHL1 and ATF1 in hpm91 is to yield various intermediates satisfying increased carbon demand under anoxic conditions (Weigelt et al. 2009). Yet, a large variation of the key enzymes in TCA cycle of hpm91, such as malate dehydrogenase MDH4 and subunits of succinate dehydrogenase (SDH1, SDH2, SDH4) was observed (Table S3). This may reflect dynamic energetic status in hpm91 during sulfur-deprived H2 production.
hpm91 is characteristic of increased photosynthetic core and decreased PSI antenna
In Table S3, nearly 50% of the PSII and PSI proteins were accumulated in hpm91 especially PsbH, PsbP, PsbQ, PsaA, PsaB, PsaD, and PsaF with average values of fold-increase within 3.1 to 25.2. Because H2 evolution profile of wild type and hpm91 was mostly distinct at 120 h of sulfur deprivation (Chen et al. 2019), the change-fold of this time point was compared between the two strains (Fig. 4b, upright pannel). Compared to wild type, fold-increase values of the three PSII proteins was about 2-times larger in hpm91. Considering that PsbP and PsbQ are essential in maintaining water-splitting reaction (Shen 2015) and PsbH is crucial for stable assembly and optimal function of PSII (Umena et al. 2011; Trosch et al. 2018), their greater increase in hpm91 may partially explain its significantly higher residual PSII activity during prolonged H2 production (Chen et al. 2019). Regarding to LHCII proteins, it was noted that decrease of LHCBM3, LHCBM6, LHCBM8 were more pronounced in hpm91 under such conditions (Fig. 4b, upright panel). Most interestingly, we found that while the fold-increase values of PSI core subunits of hpm91 were either higher (PsaB and PsaF) or comparable (PsaA, PsaD) to wild type, all the LHCA proteins displayed declined trends with greater values of decrease-fold in hpm91 than wild type during H2 production process (Fig. 4b, upright panel). These results strongly suggest that PGR5-deficient hpm91 mutant is an algal strain characteristic of small-sized PSI antenna under not only normal condition (Table 1) but also during sustained sulfur-deprived H2 production. It is generally known that, in wild type, transcriptional regulation of LHC genes plays a central role in antenna size adjustment. To test if this is true for hpm91 mutant, we then carried out qRT-PCR analysis of the genes encoding the LHCA proteins. Our data showed that their mRNA levels were indeed down-regulated during H2 production (Fig. S3). Taken together, we suggest that mutation of PGR5 caused not only the impaired CEF (Fig. S2a) but also significantly reduced PSI antenna (Fig. 4b), leading to its higher efficiency of light utilization than wild type toward H2 photoproduction (Kosourov et al. 2011).
Strikingly, we found the level of LHCA2 protein in hpm91 was remarkably decreased during H2 production, whereas no change was revealed in wild type (Fig. 4b, upright panel). This distinction could be of strong indication of LHCA2 as a negative effector on H2 photoproduction in Chlamydomonas. Indeed, a recent report by Ho et al. (Ho et al. 2022) shows that pgr5/lhca2 double mutant produced more than twofold H2 amount relative to its single pgr5 mutant, revealing the crucial role of LHCA2 involved in algal H2 photoproduction, strongly suggesting that lhca2 is a potent gene target for further genetic modifications of the organism toward H2 photoproduction.
Reinforced cell redox homeostasis and anti-oxidative systems in hpm91
Based on comparison of Tables S2 and S3, a higher percentage of up-regulated proteins involved in cell redox homeostasis and/or anti-oxidative stress responses was revealed in hpm91 than wild type under such condition. Although most of them overlapped with wild type, accumulation of several proteins in TRX superfamily, mitochondrial proteins SCO1 and cytochrome c peroxidase CCPR1 as well as those related to oxidative stress responses was more pronounced in hpm91 than wild type. Strikingly, the amount of TRXh, PDI2, APX1, and CCPR1 increased at least tenfold in hpm91 during prolonged H2 production (Fig. 4b, downleft panel). Based on physiological and biochemical analysis of wild-type cells, it has been earlier suggested that oxidative stress occurs during sulfur-deprived H2 photoproduction (Sáenz et al. 2015; Kosourov et al. 2017). Regarding the pgr5 mutants (pgr5, hpm91), we have previously observed both increased ROS tolerance and ROS-scavenging enzyme activity under such conditions (Chen et al. 2016). In this work, we found the higher percentage and abundance of those proteins involved in cell redox homeostasis and/or anti-oxidative stress reactions (Tables S2 and S3; Fig. 4b, downleft panel). Together with the finding of better cell viability of hpm91 than wild type during sulfur-deprived H2 photoproduction (Fig. 2), we suggest that the lower amount of ROS observed in hpm91 (Fig. 2c and d) could be largely attributed to the marked increase of both protein abundance of those and activity of the ROS-scavenging enzymes as well as putatively reduced PSI antenna mentioned above (Lu et al. 2021). A question is open how this is fulfilled in the mutant cells. Considering that H2 production is beneficial for cell survival and maintenance of photosynthetic apparatus activity as well as energy and redox status under such conditions (Chen et al., 2016; 2019; Antal et al. 2020), we presume that shifting to the H2 production mode is one of the best choices for the mutant cells acclimating to sulfur-deprived anaerobic condition.
Enhanced N- and S- metabolism in hpm91 during sustained H2 photoproduction
Comparison of Tables S2 and S3 reveals that both N- and S- metabolism of hpm91 was also enhanced during sustained H2 photoproduction. Considering that glutamine and glutamate are the major intracellular amino group donors for the synthesis of several other amino acids and nitrogen-containing compounds including purine and pyrimidine nucleobases (Zhang et al. 2018), we combined the functional groups of ‘amino acid pathways,’ ‘terpenoid- and de novo biosynthesis of pyrimidine nucleobase’ (Fig. 4a) and referred as N-metabolism in Table S3. In contrast to wild type, 3 proteins of those were only accumulated in hpm91 at 120 h of sulfur deprivation. These were aspartate aminotransferase AST3, glutamate dehydrogenase GDH and GDH2, showing upto 2.20-, 13.42-, and 24.56-fold increase (Table S3). AST3 is known to be one of the major enzymes catalyzing conversion of glutamate and oxaloacetic acid (OAA) into asparate and 2-oxoglutarate (2-OG/alpha-KG), an intermediate of the TCA cycle (Ohashi et al. 2011) that serves as the metabolic basis for coupling N- and C-metabolisms in photosynthetic organisms. Because AST3 was already higher-expressed in hpm91 under normal condition (Table 1), the continued increase is strongly indicating its dominant role in asparate biosynthesis and/or maintenance of C/N metabolic balance in the PGR5-deficient hpm91 mutant during prolonged H2 production.
More interestingly, 2 glutamate dehydrogenases (GDH and GDH2) were markedly accumulated in hpm91 during sustained H2 production. These proteins are supposed to play an anaplerotic role in ammonium assimilation via conversion of Glu into 2-OG and ammonium in Chlamydomonas (Moyano et al. 1995). Their remarkable increase in hpm91 implicates activation of this minor pathway of ammonium assimilation in the mutant under such condition. Together with upregulation of NADH-dependent glutamate synthase GSN1 (Table S3), the key enzyme of the major route GS-GOGAT cycle, we propose that, due to loss of PGR5, both the major and minor route of ammonium assimilation was activated/or enhanced in hpm91 during sustained H2 photoproduction. Because enhanced ammonium assimilation requires higher demand of the carbon skeleton 2-OG (Ohashi et al. 2011) for coupling between nitrogen and carbon metabolism (Zhang et al. 2018), we propose that by stimulating the anaplerotic role of the GDH toward producing more 2-OG, carbon, and nitrogen metabolism could be better coupled in hpm91 than wild type under such conditions.
In this work, we also found remarkable accumulations for 5 key proteins involved in sulfur assimilation in hpm91 during H2 photoproduction (Table S3). These include not only the above-mentioned APR1 (Table 1) with continued increase in abundance (Table S3) but also ATP-sulfurylases (ATS1, ATS2) as well as cysteine synthase OASTL3 toward incorporation SO42+ into cysteine (Gonzales-Ballester et al. 2009). Because the latter two proteins were found lower-expressed in hpm91 under normal condition (Table 1), their marked accumulation and that of ATS1 during prolonged H2 production may strongly indicate activation of the pathway toward cysteine biosynthesis (Fig. 4b, downright pannel). Meanwhile, we observed upto 4.5-fold accumulation of APK1 in hpm91 during H2 production process (Table S3), suggesting sulfur-assimilation pathway toward cysteine biosynthesis sulfation of metabolites cysteine biosynthesis (Gonzales-Ballester et al. 2009) was also enhanced in hpm91 (Fig. 4b, downright panel). These results strongly indicate that overall sulfur metabolism was more enhanced in hpm91 relative to wild type during sulfur-deprived H2 production process.
Creating mutants with H2 production excess to hpm91
Since the data described above suggests that hpm91 is better suited for sulfur-deprived H2 production photosynthetically, metabolically, and redox poised, we hypothesized that the strain can be used as a “chassis cell” for creating new mutant strains with enhanced H2 production relative to hpm91. To test this, an insertion mutant library derived from hpm91 was constructed according to (Kindle 1990; Zhao et al. 2017) followed by tranformants selection via zeocin resistance (Fig. 5a). Considering that increased stability of PSII is essential to sustain sufur-deprived H2 photoproduction in Chlamydomonas (Volgusheva et al. 2013; Chen et al. 2019), a subsequent ‘two-step mutant screening’ was applied, i. e. Y(II) measurements using Maxi-Imaging PAM chlorophyll fluorometer (Walz, Germany) as the first followed by H2-generating phenotype confirmation by GC analysis as described (Sun et al. 2013). Preliminary mutant screening identified over two hundred transformants with 10% increase of Y(II) values than hpm91 under sulfur deprivation. Subsequent screening by GC analysis identified one of the mutants, named hpm91-108, with significantly higher H2 production than hpm91 (Fig. 5b). At 120 h, H2 production in hpm91-108 was 55. 9% higher than hpm91. This result is the direct experimental evidence of hpm91 as a potent strain for re-engineering the organism toward advancing photobiological H2 production.
Screening for H2 production mutants excess to hpm91 a Outline of the screening method. b Phenotype confirmation of the hpm91-derived mutants. H2-producing capability of the selected mutants was determined with a GC-2014 gas chromatographer (Shimadzu; Japan) at 5 days of sulfur deprivation. Standard deviations were estimated from 3 biological replicates
Concluding remarks
In summary, scaling-up and in-depth analysis of hpm91 mutant has revealed several valuable properties toward development of sunlight-powered algal H2 production systems in the near future. First, it is largely up-scalable using the ‘two-step’ protocol of H2 induction by sulfur deprivation (Melis et al. 2000). In both steps, up to 100-fold extension of PBR (10 L, mixotrophic growth) (Chen et al. 2019) and HPBR (10 L, H2 photoproduction) was achieved in the laboratory set-ups, leading to an average H2 output of 7287 ml/10L-HPBR for averagely 26 days (this work). Second, hpm91 is robust during prolonged H2 production. In the absence of PGR5, hpm91 shows competent viability than wild type and remains active over a long period of sulfur deprivation, which could be mainly due to a decrease of intracellular ROS involved in signaling pathways (Mullineaus et al. 2008). Third, hpm91 was active metabolically (reinforced anti-ROS systems, compromised carbon metabolism, enhanced/activation of anaplerotic route of ammonium assimilation and sulfur assimilation) and photosynthetically (optimal structure and function of PSII and PSI, reduced size of PSI antenna and CEF) toward sustained H2 photoproduction. These results reveal not only new insights of cellular and molecular basis of enhanced H2 production in hpm91 but also provide additional candidate gene targets and modules for further genetic modifications and/or in artificial photosynthesis mimics (Ye et al. 2021) toward basic and applied research aiming at advancing solar-H2 technology.
References
Antal T, Krendeleva TE, Laurinavichene TV, Makarova VV, Ghirardi ML, Rubin AB, Tsygankov AA, Seibert M (2009) Relationships between H2 photoproduction and different electron transport pathways in sulfur-deprived Chlamydomonas reinhardtii. Int J Hydrog Energ 34:9087–9094. https://doi.org/10.1016/j.bbabio.2003.09.008
Antal T, Petrova E, Slepnyova V, Kukarskikh G, Volgusheva A, Dubini A, Baizhumanov A, Taina Tyystjärvi T et al (2020) Photosynthetic hydrogen production as acclimation mechanism in nutrient-deprived Chlamydomonas. Algal Res 40:101951. https://doi.org/10.1016/j.algal.2020.101951
Arnon DI (1949) Copper enzymes in isolated chloroplasts. polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15. https://doi.org/10.1104/pp.24.1.1
Bayro-Kaiser V, Nelson N (2017) Microalgal hydrogen production: prospects of an essential technology for a clean and sustainable energy economy. Photosynth Res 133:49–62. https://doi.org/10.1007/s11120-017-0350-6
Chen M, Zhao K, Sun YL, Cui SX, Zhang LF, Yang B, Wang J, Kuang TY, Huang F (2010) Proteomic analysis of hydrogen photoproduction in sulfur-deprived Chlamydomonas cells. J Proteome Res 9:3854–3866. https://doi.org/10.1021/pr100076c
Chen M, Zhang J, Zhao L, Xing JL, Peng L, Kuang T, Rochaix JD, Huang F (2016) Loss of algal proton gradient regulation 5 increases reactive oxygen species scavenging and H2 evolution. J Integr Plant Biol 58:943–946. https://doi.org/10.1111/jipb.12502
Chen M, Liu P, Zhang F, Peng LW, Huang F (2019) Photochemical characteristics of Chlamydomonas mutant hpm91 lacking proton gradient regulation 5 (PGR5) during sustained H2 photoproduction under sulfur deprivation. Int J Hydrogen Energ 44:31790–31799. https://doi.org/10.1016/j.ijhydene.2019.10.074
Chochois V, Dauvillee D, Beyly A, Tolleter D, Cuine S, Timpano H, Ball S, Cournac L, Peltier G (2009) Hydrogen production in Chlamydomonas: photosystem II-dependent and -independent pathways differ in their requirement for starch metabolism. Plant Physiol 151:631–640. https://doi.org/10.1104/pp.109.144576
Forestier M, King P, Zhang LP, Posewitz M, Schwarzer S, Happe T, Ghirardi ML, Seibert M (2003) Expression of two [Fe]-hydrogenases in Chlamydomonas reinhardtii under anaerobic conditions. Eur J Biochem 270:2750–2758. https://doi.org/10.1046/j.1432-1033.2003.03656
Fouchard S, Hemschemeier A, Caruana A, Pruvost J, Legrand J, Happe T, Peltier G, Cournac L (2005) Autotrophic and mixotrophic hydrogen photoproduction in sulfur-deprived chlamydomonas cells. Appl Environ Microbiol 71:6199–6205. https://doi.org/10.1128/AEM.71.10.6199-6205.2005
Ge HT, Fang LF, Huang XH, Wang JL, Chen WY, Liu Y, Zhang YY, Wang X et al (2017) Translating divergent environmental stresses into a common proteome response through the histidine kinase 33 (Hik33) in a model Cyanobacterium. Mol Cell Proteomics 16:1258–1274. https://doi.org/10.1074/mcp.M116.068080
Ghirardi ML (2015) Implementation of photobiological H2 production: the O2 sensitivity of hydrogenases. Photosynth Res 125:383–393. https://doi.org/10.1007/s11120-015-0158-1
Gonzales-Ballester D, Grossman AR (2009) Sulfur: From acquisition to assimilation. In: DB Stern, (ed) The Chlamydomonas Sourcebook, Second Edition, Elsevier, https://doi.org/10.1016/B978-0-12-370873-1.00013-7.
Gorman DS, Levine RP (1965) Cytochrome f and plastocyanin - their sequence in photosynthetic electron transport chain of Chlamydomonas reinhardi. Proc Natl Acad Sci USA 54:1665–1669. https://doi.org/10.1073/pnas.54.6.1665
Gutierrez-Marcos JF, Roberts MA, Campbell EI, Wray JL (1996) Three members of a novel small gene-family from Arabidopsis thaliana able to complement functionally an Escherichia coli mutant defective in PAPS reductase activity encode proteins with a thioredoxin-like domain and “APS reductase” activity. Proc Natl Acad Sci USA 93:13377–13382. https://doi.org/10.1073/pnas.93.23.13377
Hemschemeier A, Melis A, Happe T (2009) Analytical approaches to photobiological hydrogen production in unicellular green algae. Photosynth Res 102:523–540. https://doi.org/10.1007/s11120-009-9415-5
Ho TTH, Schwier C, Elman T, Fleuter V, Zinzius K, Scholz M, Yacoby I, Buchert F, Hippler M (2022) Photosystem I light-harvesting proteins regulate photosynthetic electron transfer and hydrogen production. Plant Physiol. https://doi.org/10.1093/plphys/kiac055
Iwai M, Takizawa K, Tokutsu R, Okamuro A, Takahashi Y, Minagawa J (2010) Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature 464:1210–1213. https://doi.org/10.1038/nature08885
Johnson X, Steinbeck J, Dent RM, Takahashi H, Richaud P, Ozawa SI, Houille-Vernes L, Petroutsos D et al (2014) Proton gradient regulation 5-mediated cyclic electron flow under ATP- or redox-limited conditions: A study of ∆ATPase pgr5 and ∆rbcL pgr5 mutants in the green alga Chlamydomonas reinhardtii. Plant Physiol 165:438–452. https://doi.org/10.1104/pp.113.233593
Kindle KL (1990) High-Frequency nuclear transformation of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 87:1228–1232. https://doi.org/10.1073/pnas.87.3.1228
Kosourov S, Tsygankov A, Seibert M, Ghirardi ML (2002) Sustained hydrogen photoproduction by Chlamydomonas reinhardtii: Effects of culture parameters. Biotechnol Bioeng 78:731–740. https://doi.org/10.1002/bit.10254
Kosourov SN, Ghirardi ML, Seibert M (2011) A truncated antenna mutant of Chlamydomonas reinhardtii can produce more hydrogen than the parental strain. Int J Hydrogen Energ 36:2044–2048. https://doi.org/10.1016/j.ijhydene.2010.10.041
Kosourov S, Murukesan G, Seibert M, Allahverdiyeva Y (2017) Evaluation of light energy to H2 energy conversion efficiency in thin films of cyanobacteria and green alga under photoautotrophic conditions. Algal Res 28:253–263. https://doi.org/10.1016/j.algal.2017.09.027
Kruse O, Rupprecht J, Bader KP, Thomas-Hall S, Schenk PM, Finazzi G, Hankamer B (2005) Improved photobiological H2 production in engineered green algal cells. J Biol Chem 280:34170–34177. https://doi.org/10.1074/jbc.M503840200
Lu Y, Gan Q, Iwai M, Alboresi A, Burlacot A, Dautermann O, Takahashi H, Crisanto T et al (2021) Role of an ancient light-harvesting protein of PSI in light absorption and photoprotection. Nat Commun 12:679. https://doi.org/10.1038/s41467-021-20967-1
Matthew T, Zhou W, Rupprecht J, Lim L, Thomas-Hall SR, Doebbe A, Kruse O, Hankamer B et al (2009) The metabolome of Chlamydomonas reinhardtii following induction of anaerobic H2 production by sulfur depletion. J Biol Chem 284:23415–23425. https://doi.org/10.1074/jbc.M109.003541
Melis A, Zhang LP, Forestier M, Ghirardi ML, Seibert M (2000) Sustained photobiological hydrogen gas production upon reversible inactivation of oxygen evolution in the green alga Chlamydomonas reinhardtii. Plant Physiol 122:127–135. https://doi.org/10.1104/pp.122.1.127
Merchant SS, Prochnik SE, Vallon O, Harris EH, Karpowicz SJ, Witman GB, Terry A, Salamov A et al (2007) The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318:245–251. https://doi.org/10.1126/science.1143609
Moyano E, Cardenas J, Munozblanco J (1995) Involvement of NAD(P)+- glutamate dehydrogenase isoenzymes in carbon and nitrogen metabolism in Chlamydomonas reinhardtii. Physiol Plantarum 94:553–559. https://doi.org/10.1034/j.1399-3054.1995.940403.x
Mullineaux PM, Karpinski S, Creissen GP (2008) Integration of Signaling in Antioxidant Defenses. In Demmig-Adams, B, Adams WW, Mattoo AK (eds) Photoprotection, Photoinhibition, Gene Regulation, and Environment Advances in Photosynthesis and Respiration, Springer, Dordrecht 223–239 https://doi.org/10.1007/1-4020-3579-9_15.
Munekage Y, Hojo M, Meurer J, Endo T, Tasaka M, Shikanai T (2002) PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 110:361–371. https://doi.org/10.1016/S0092-8674(02)00867-X
Nguyen AV, Thomas-Hall SR, Malnoe A, Timmins M, Mussgnug JH, Rupprecht J, Kruse O, Hankamer B, Schenk PM (2008) Transcriptome for photobiological hydrogen production induced by sulfur deprivation in the green alga Chlamydomonas reinhardtii. Eukaryot Cell 7:1965–1979. https://doi.org/10.1128/EC.00418-07
Nishiyama H, Yamada T, Nakabayashi M, Maehara Y, Yamaguchi M, Kuromiya Y, Nagatsuma Y, Tokudome H et al (2021) Photocatalytic solar hydrogen production from water on a 100–m2 scale. Nature 598:304–307. https://doi.org/10.1038/s41586-021-03907-3
Ohashi Y, Shi W, Takatani N, Aichi M, Maeda S, Watanabe S, Yoshikawa H, Omata T (2011) Regulation of nitrate assimilation in cyanobacteria. J Exp Bot 62:1411–1424. https://doi.org/10.1093/jxb/erq427
Philipps G, Krawietz D, Hemschemeier A, Happe T (2011) A pyruvate formate lyase-deficient Chlamydomonas reinhardtii strain provides evidence for a link between fermentation and hydrogen production in green algae. Plant J 66:330–340. https://doi.org/10.1111/j.1365-313X.2011.04494.x
Ravina CG, Chang CI, Tsakraklides GP, McDermott JP, Vega JM, Leustek T, Gotor C, Davies JP (2002) The sac mutants of Chlamydomonas reinhardtii reveal transcriptional and posttranscriptional control of cysteine biosynthesis. Plant Physiol 130:2076–2084. https://doi.org/10.1104/pp.012484
Sáenz ME, Bišová K, Touloupakis E, Faraloni C, Marzio WDD, Torzillo G (2015) Evidences of oxidative stress during hydrogen photoproduction in sulfur-deprived cultures of Chlamydomonas reinhardtii. Int J Hydrogen Energ 40:10410–10417. https://doi.org/10.1016/j.ijhydene.2015.06.124
Schonfeld C, Wobbe L, Borgstadt R, Kienast A, Nixon PJ, Kruse O (2004) The nucleus-encoded protein MOC1 is essential for mitochondrial light acclimation in Chlamydomonas reinhardtii. J Biol Chem 279:50366–50374. https://doi.org/10.1074/jbc.M408477200
Schwenkert S, Fernie AR, Geigenberger P, Leister D, Mohlmann T, Naranjo B, Neuhaus HE (2022) Chloroplasts are key players to cope with light and temperature stress. Trends Plant Sci. https://doi.org/10.1016/j.tplants.2021.12.004
Scoma A, Krawietz D, Faraloni C, Giannelli L, Happe T, Torzillo G (2012) Sustained H2 production in a Chlamydomonas reinhardtii D1 protein mutant. J Biotechnol 157:613–619. https://doi.org/10.1016/j.jbiotec.2011.06.019
Setya A, Murillo M, Leustek T (1996) Sulfate reduction in higher plants: Molecular evidence for a novel 5’-adenylylsulfate reductase. Proc Natl Acad Sci USA 93:13383–13388. https://doi.org/10.1073/pnas.93.23.13383
Shen JR (2015) The structure of Photosystem II and the mechanism of water oxidation in photosynthesis. Annu Rev Plant Biol 66:23–48. https://doi.org/10.1146/annurev-arplant-050312-120129
Shen LL, Huang ZH, Chang SH, Wang WD, Wang JF, Kuang TY, Han GY, Shen JR, Zhang X (2019) Structure of a C2S2M2N2 type PSII-LHCII supercomplex from the green alga Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 116:21246–21255. https://doi.org/10.1073/pnas.1912462116
Steinbeck J, Nikolova D, Weingarten R, Johnson X, Richaud P, Peltier G, Hermann M, Magneschi L, Hippler M (2015) Deletion of proton gradient regulation 5 (PGR5) and PGR5-like 1 (PGRL1) proteins promote sustainable light-driven hydrogen production in Chlamydomonas reinhardtii due to increased PSII activity under sulfur deprivation. Front Plant Sci 6:892. https://doi.org/10.3389/fpls.2015.00892
Su XD, Ma J, Pan XW, Zhao XL, Chang WR, Liu ZF, Zhang XZ, Li M (2019) Antenna arrangement and energy transfer pathways of a green algal photosystem-I-LHCI supercomplex. Nat Plants 5:273–281. https://doi.org/10.1038/s41477-019-0380-5
Suga M, Ozawa SI, Yoshida-Motomura K, Akita F, Miyazaki N, Takahashi Y (2019) Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I. Nat Plants 5:626–636. https://doi.org/10.1038/s41477-019-0438-4
Sun YL, Chen M, Yang HM, Zhang J, Kuang TY, Huang F (2013) Enhanced H2 photoproduction by down-regulation of ferredoxin-NADP+ reductase (FNR) in the green alga Chlamydomonas reinhardtii. Int J Hydrogen Energ 38:16029–16037. https://doi.org/10.1016/j.ijhydene.2013.10.011
Suorsa M, Jarvi S, Grieco M, Nurmi M, Pietrzykowska M, Rantala M, Kangasjarvi S, Paakkarinen V et al (2012) Proton gradient regulation5 is essential for proper acclimation of arabidopsis photosystem I to naturally and artificially fluctuating light conditions. Plant Cell 24:2934–2948. https://doi.org/10.1105/tpc.112.097162
Takahashi H, Clowez S, Wollman FA, Vallon O, Rappaport F (2013) Cyclic electron flow is redox-controlled but independent of state transition. Nat Commun 4:1954. https://doi.org/10.1038/ncomms2954
Toepel J, Illmer-Kephalides M, Jaenicke S, Straube J, May P, Goesmann A, Kruse O (2013) New insights into Chlamydomonas reinhardtii hydrogen production processes by combined microarray/RNA-seq transcriptomics. Plant Biotechnol J 11:717–733. https://doi.org/10.1111/pbi.12062
Tolleter D, Ghysels B, Alric J, Petroutsos D, Tolstygina I, Krawietz D, Happe T, Auroy P et al (2011) Control of hydrogen photoproduction by the proton gradient generated by cyclic electron flow in Chlamydomonas reinhardtii. Plant Cell 23:2619–2630. https://doi.org/10.1105/tpc.111.086876
Trosch R, Barahimipour R, Gao Y, Badillo-Corona JA, Gotsmann VL, Zimmer D, Muhlhaus T, Zoschke R, Willmund F (2018) Commonalities and differences of chloroplast translation in a green alga and land plants. Nat Plants 4:564–575. https://doi.org/10.1038/s41477-018-0211-0
Udeshi ND, Svinkina T, Mertins P, Kuhn E, Mani DR, Qiao JW, Carr SA (2013) Refined preparation and use of anti-diglycine remnant (K-epsilon-GG) antibody enables routine quantification of 10,000s of ubiquitination sites in single proteomics experiments. Mol Cell Proteomics 12:825–831. https://doi.org/10.1074/mcp.O112.027094
Umena Y, Kawakami K, Shen JR, Kamiya N (2011) Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9A. Nature 473:55–60. https://doi.org/10.1038/nature09913
Volgusheva A, Styring S, Mamedov F (2013) Increased photosystem II stability promotes H2 production in sulfur-deprived Chlamydomonas reinhardtii. Proc Natl Acad Sci USA 110:7223–7228. https://doi.org/10.1073/pnas.1220645110
Wang H, Alvarez S, Hicks LM (2012) Comprehensive comparison of iTRAQ and label-free LC-based quantitative proteomics approaches using two Chlamydomonas reinhardtii strains of interest for biofuels engineering. J Proteome Res 11:487–501. https://doi.org/10.1021/pr2008225
Weigelt K, Kuster H, Rutten T, Fait A, Fernie AR, Miersch O, Wasternack C, Emery RJN et al (2009) ADP-glucose pyrophosphorylase-deficient pea embryos reveal specific transcriptional and metabolic changes of carbon-nitrogen metabolism and stress responses. Plant Physiol 149:395–411. https://doi.org/10.1104/pp.108.129940
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. https://doi.org/10.1038/nmeth.1322
Yacoby I, Pochekailov S, Toporik H, Ghirardi ML, King PW, Zhang SG (2011) Photosynthetic electron partitioning between [FeFe]-hydrogenase and ferredoxin:NADP+-oxidoreductase (FNR) enzymes in vitro. Proc Natl Acad Sci USA 108:9396–9401. https://doi.org/10.1073/pnas.1103659108
Yang WQ, Catalanotti C, Wittkopp TM, Posewitz MC, Grossman AR (2015) Algae after dark: mechanisms to cope with anoxic/hypoxic conditions. Plant J 82:481–503. https://doi.org/10.1111/tpj.12823
Ye S, Shi WW, Liu Y, Li DF, Yin H, Chi HB, Luo YL, Ta N et al (2021) Unassisted photoelectrochemical cell with multimediator modulation for solar water splitting exceeding 4% solar-to-hydrogen efficiency. J Am Chem Soc 143:12499–12508. https://doi.org/10.1021/jacs.1c00802
Zhang LP, Happe T, Melis A (2002) Biochemical and morphological characterization of sulfur-deprived and H2-producing Chlamydomonas reinhardtii (green alga). Planta 214:552–561. https://doi.org/10.1007/s004250100660
Zhang ZD, Shrager J, Jain M, Chang CW, Vallon O, Grossman AR (2004) Insights into the survival of Chlamydomonas reinhardtii during sulfur starvation based on microarray analysis of gene expression. Eukaryot Cell 3:1331–1348. https://doi.org/10.1128/Ec.3.5.1331-1348.2004
Zhang CC, Zhou CZ, Burnap RL, Peng L (2018) Carbon/Nitrogen metabolic balance: lessons from Cyanobacteria. Trends Plant Sci 23:1116–1130. https://doi.org/10.1016/j.tplants.2018.09.008
Zhao L, Chen M, Cheng DM, Yang HM, Sun YL, Zhou HY, Huang F (2013) Different B-type methionine sulfoxide reductases in Chlamydomonas may protect the alga against high-light, sulfur-depletion, or oxidative stress. J Integr Plant Biol 55:1054–1068. https://doi.org/10.1111/jipb.12104
Zhao L, Cheng DM, Huang XH, Chen M, Dall’Osto L, Xing JL, Gao LY, Li LY et al (2017) A light harvesting complex-like protein in maintenance of photosynthetic components in Chlamydomonas. Plant Physiol 174:2419–2433. https://doi.org/10.1104/pp.16.01465
Acknowledgements
This work was supported by funding from the National Natural Science Foundation of China (31470340), the Ministry of Science and Technology of China (2015CB150100) and CAS (XDB17030300, KGCX2-YW-373).
Author information
Authors and Affiliations
Contributions
MC, YW and FH conceived the project and designed the research; PL, DY, MC, JZ, LS and XH carried out the experiments and data analysis; XX, KX, YY and YG performed genomic sequencing and data analysis; PL and DY prepared data files; PL, DY, YW and FH wrote the manuscript, with input from all authors. All authors approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests. This work is included in a PCT patent application (PCT/CN2022/099056) by Institute of Botany, Chinese Academy of Sciences.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
11120_2022_945_MOESM1_ESM.docx
Supplementary file1 Fig. S1 Comparison of percentage of dead cells in the culture of hpm91 and wild type during 120 h of sulfur-deprived H2 production. Fig. S2 Functional and genomic verification of PGR5 deletion in hpm91. Fig. S3 qRT-PCR analysis of relative gene expression of the LHCAs in hpm91 during 120 h of sulfur-deprived H2 production (DOCX 1962 kb)
11120_2022_945_MOESM2_ESM.docx
Supplementary file2 Table S1 Comparison of H2 output of hpm91 in 10L-HPBR under different light intensities. Table S2 List of differentially expressed proteins in wild type delineated from Dataset 5 that represents major proteome changes during H2 production process. Table S3 List of differentially expressed proteins in hpm91 delineated from Dataset 6 that represents major proteome changes during H2 production process. Table S4 List of primers used in this work (DOCX 219 kb)
Supplementary file3 Demonstration of a laboratory set-up making a H2-fuel-cell-powered toycar drive by input of algal-H2 collected from hpm91 and ambient air. (AVI 2917 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, P., Ye, DM., Chen, M. et al. Scaling-up and proteomic analysis reveals photosynthetic and metabolic insights toward prolonged H2 photoproduction in Chlamydomonas hpm91 mutant lacking proton gradient regulation 5 (PGR5). Photosynth Res 154, 397–411 (2022). https://doi.org/10.1007/s11120-022-00945-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11120-022-00945-4