Abstract
Rice (Oryza sativa) is a tropical cereal crop that is severely affected by chilling stress at the seedling stage, although glutinous rice 89-1 (Gr89-1) in Chongqing, China, shows tolerance to low temperatures and overwintering ability. However, little research has been conducted on the mechanisms regulating chilling stress in Gr89-1. In this study, a comprehensive of transcriptional profiles of Gr89-1 seedlings at the three-leaf stage was conducted after a 4 °C treatment for 2, 6, 12, 24, or 48 h. Overall, 2993 differentially expressed genes were detected in Gr89-1 seedlings upon cold exposure. Gene Ontology testing and pathway analysis revealed differentially expressed genes involved in transcriptional regulation, carbohydrate metabolism, plant hormone signal, and cell wall composition. A total of 243 transcription factors were differentially expressed during the cold treatment; in particular, the AP2/EREBP, bHLH, NAC, WRKY, C2H2, and TIFY families were generally upregulated after cold treatment, whereas the mTERF and GNAT families were downregulated. Chilling stress changed the starch and sucrose metabolism, coupled with the accumulation of sucrose and trehalose level, and increases in jasmonic acid level in Gr89-1 seedlings. Furthermore, a number of the cell wall-related genes identified in the present study were also differentially expressed during the cold treatment. The genes and pathways identified in the current study increase our understanding of the mechanisms underlying cold resistance in rice seedlings.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Chilling (0–15 °C) stress severely affects rice growth and development in temperate and sub-tropical areas. Rice seedlings during early spring in such areas are often subjected to low temperatures, leading to poor growth and reduced yield (Baruah et al. 2009). Most temperate plants have evolved regulatory mechanisms to adapt to unfavorable low-temperature environments through a complex process known as cold acclimation (Thomashow 1999). The C-repeat binding factor/dehydration-responsive element-binding protein 1 (CBF/DREB1; DREB1A, DREB1B, and DREB1C)-dependent response pathway plays a vital role in cold tolerance during cold acclimation (Chinnusamy et al. 2007; Kim 2007; Shinozaki et al. 2003). CBFs mediate cold-responsive genes with various functions, including transcription, osmoprotectant biosynthesis, and signal transduction events (e.g., plant hormone signaling), among many others (Krasensky et al. 2012; Lissarre et al. 2010).
The amount of compatible solutes (e.g., amino acids, amines, and carbohydrates) is particularly important in the stabilization of membranes and associated with the chilling tolerance (Krasensky and Jonak 2012). Many studies have found that soluble carbohydrate, sucrose, trehalose, and stachyose drastically accumulated in plants under low temperatures (Krasensky and Jonak 2012). Phytohormones regulate multiple aspects of plant growth and development, and also play substantial direct or indirect roles in plant responses to diverse stressors (Jeon and Kim 2013). Jasmonic acid (JA) represents an important class of lipid-derived plant hormones involved in the cold response (Peleg and Blumwald 2011). Endogenous free JA accumulates during low-temperature conditions in camellias and wheat (Li et al. 2016; Qian et al. 2016). Methyl jasmonate treatment increases the chilling tolerance of banana, pomegranate fruit, and wheat seedlings (Qi et al. 2006; Zhao et al. 2013; Zolfagharinasab and Hadian 2007). The catalytic steps in JA biosynthesis involving the conversion of α-linolenic acid to JA are mediated by the action of enzymes encoded by the lipoxygenase (LOX), allene oxide synthase (AOS), allene oxide cyclase (AOC), 12-oxophytodienoic acid reductase (OPR), and β-oxidase genes (Wasternack and Hause 2013). After synthesis, the COI1-JAZ co-receptor perceives bioactive JA-Ile. As a negative regulator of the JA signaling pathway, the JASMONATE-ZIM-DOMAIN (JAZ) protein regulates the expression of JA-responsive genes by inhibiting the activities of a series of downstream transcription factors (TFs) (Hu et al. 2017). JA acts as a signal in the ICE-CBF pathway, which is involved in cold tolerance in Arabidopsis and tomato (Hu et al. 2013; Wang et al. 2016). However, the integrative regulatory effects of JA in rice subject to cold are poorly understood.
Transcriptomic analyses have provided new insight into the complex stress response mechanisms of plants. A large number of genome-wide gene profiling studies on rice responding to low temperature have been conducted. Most of the cold-responsive genes identified are related to transcription regulation, osmoprotectant accumulation, detoxification, ROS scavenging, primary energy metabolism, cell wall organization, and hormone-related signal transduction processes (Buti et al. 2018; Chawade et al. 2013; Pradhan et al. 2019; Zhang et al. 2012a, b; Zhang et al. 2017a, b). Glutinous rice 89-1 (Gr89-1) has overwintering ability and germinates through axillary buds during the following spring. Yields are the same as in a normal season (up to 6.29 t/hm2) in Chongqing, China (Deng et al. 2018). Other studies have revealed similar overwintering ability of Oryza longistaminata and Dongxiang wild rice (O. rufipogon) in south and southwest China (He et al. 1996; Liang et al. 2017; Mao et al. 2015; Zhang et al. 2017a, b). Genome-wide gene profiling has shown that genes associated with CBF/DREB1 regulon transcriptional regulation are significantly upregulated in wild rice (O. longistaminata) shoots in response to long-term low temperatures (Zhang et al. 2017a, b). In a previous study, Gr89-1 had better seedling-stage chilling tolerance according to a chlorophyll fluorescence analysis (Wang et al. 2012). Gr89-1 has clearly developed methods to protect itself against chilling stress, and genome-wide gene profiling provides a valuable method for understanding the molecular mechanisms underlying the low temperature response. In the present study, we analyzed transcriptomic profile changes in Gr89-1 seedlings exposed to a cold treatment (4 °C for 2, 6, 12, 24, or 48 h) using a deep high-throughput sequencing-based digital gene expression analysis. Gene Ontology (GO) testing, and combined biological pathway and physiological data analysis, indicated that Gr89-1 improves its response to chilling stress by increasing the production of osmoprotectants (soluble sugar, sucrose, and trehalose), modifying its cell wall structure and composition, and altering the JA biosynthetic and signal transduction pathways.
Methods
Plant Growth and Experimental Treatment
The rice (Oryza sativa, ssp. japonica) cultivar ‘Gr89-1’, which was originally introduced by local farmers in Zhongxian County and domesticated at the Chongqing Academy of Agricultural Sciences, was used in this study. The seeds of Gr89-1 and WH86 (O. sativa, ssp. indica) were soaked in water for 24 h at room temperature, and then transplanted to plastic plates with Yoshida’s culture solution (Yoshida et al. 1972) in a growth chamber (3 M, Neuss, Germany) with a daily temperature of 26 °C and a long-day conditions (14 h light/10 h dark). The three-leaf seedlings on plates were transferred to the growth chamber under 24-h illuminations and maintained at 4 ± 1 °C for the cold treatment. Leaf samples were collected from 25 seedlings at each time point (0, 2, 6, 12, 24, and 48 h) and repeated three times. The prepared samples were frozen in liquid nitrogen and stored at − 80 °C.
RNA Isolation, Library Construction, and Bioinformatics Analysis
Total RNA was extracted using TRIzol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) and DNA was removed using RNase-free DNase I (Roche, Indianapolis, IN, USA). The Nanodrop ND-1000 (Thermo Scientific, Waltham, MA, USA) and Agilent 2100 instruments (Agilent Technologies, Palo Alto, CA, USA) were used to detect RNA quality and concentration, respectively.
Two biological replicates of the Gr89-1 seedlings sampled at the six time points were sequenced to generate 12 sequencing samples, and sequencing was performed by BGI-Shenzhen on the Illumina HiSeq™ 2000 platform, generating a total of million paired-end reads of bp length. Prior to assembly, the low-quality reads were removed. After that, a total of G bp of cleaned and paired-end reads was produced. All the sequencing saturation data are shown in Supplementary Table S1. The raw sequence data have been submitted to the NCBI Short Read Archive (SRA) with accession number PRJNA558598.
We aligned the 125-bp paired-end reads to the rice reference genome provided by the Rice Gene Annotation Project (http://rice.plantbiology.msu.edu/) at Michigan State University using TopHat (v2.0.6) (Ouyang et al. 2007; Trapnell et al. 2009), and then, unique transcript sequences of each sample were assembled using the Cuffdiff tool in the Cufflinks package (http://cole-trapnell-lab.github.io/cufflinks/) (Trapnell et al. 2012). Then, the assembled transcripts from each sample were compared with the reference annotation to reconstruct a non-redundant transcript data set using Cuffcompare. After the final transcriptome was generated, Cuffdiff was used to estimate the expression levels of all transcripts (Romualdi et al. 2003). Up- or downregulated genes under cold stress were selected with ∣log2Ratio∣ ≥ 2, p value < 0.05, and FDR < 0.001. A GO analysis was performed, with agriGO (http://bioinfo.cau.edu.cn/agriGO/index.php) used to identify the significant terms (q ≤ 0.05). Also, a metabolic pathway analysis was conducted using the Kyoto Encyclopedia of Genes and Genomes (KEGG; http://www.genome.jp/kegg/pathway.html) database.
qRT-PCR Analysis
To validate the transcriptomic data, we reverse-transcribed 2 mg total RNA per sample into first-strand cDNA with M-MLV reverse transcriptase (Promega, Madison WI, USA) and performed quantitative real-time polymerase chain reaction (qRT-PCR) on the BIO-RADCFX96 Real Time System (Bio-Rad Laboratories, Hercules, CA, USA) using a SYBR green PCR kit (Code: DRR041A; TaKaRa Bio, Shiga, Japan). Each sample had three technical replicates. To quantify the relative transcript levels of each gene, we applied the cycle threshold (ΔΔCT) method and referred to the transcript level of OsAct1 (LOC_Os05g36290). The primer sequences are listed in Supplementary Table S2.
Measurement of Chlorophyll Fluorescence
We used MONITORING-PAM (Heinz Walz GmbH) to measure chlorophyll fluorescence and estimated the photosystem II efficiencies Fv/Fm = (Fm-F0)/Fm. Twenty-five seedlings each from Gr89-1 and WH86 treated with chilling stress for 48 h were sent to the fluorometer for records at 0, 2, 6, 12, 24, and 48 h, with three replicates for each sample. Before measurement, plants went through a 1-h dark acclimation. All measurements were conducted at 10 pm.
Chemical Substance Assays
The JA contents of the seedlings at 0, 2, 6, 12, 24, and 48 h under the 4 °C condition were determined following the Liu’s procedure (Liu et al. 2018). The prepared samples were analyzed using a 4000 Q-Trap high-performance liquid chromatography-mass spectrometry (HPLC-MS) system (AB SCIEX, Framingham, MA, USA), and 10-dihydro-JA (Olchemin) for JA was used as internal standards. The contents of free proline, total soluble sugar, sucrose, and trehalose in the seedlings subjected to cold stress were determined using kits from Nanjing Jiancheng Bioengineering Institute (Nanjing, China). All experiments included three biological repeats.
Results and Discussion
Physiological Indices Indicate that Gr89-1 Exhibits Higher Cold Tolerance than WH86
To verify that Gr89-1 possesses higher cold tolerance than WH86, we treated three-leaf seedlings at 4 °C for 48 h, followed by a 1-week recovery period under normal growth conditions. Wilting and death occurred in all WH86 plants, while the recovery rate of Gr89-1 plants was 80% (Fig. 1). Prior to cold exposure, there was no difference in the Fv/Fm ratios (0.76 and 0.758) of Gr89-1 and WH86. After 24 h of cold treatment, the Fv/Fm ratio dropped to 0.12 in WH86 and to 0.622 in Gr89-1. After 48 h, the ratio was almost zero in WH86, versus 0.456 in Gr89-1 (Fig. 1c). These results demonstrate that Gr89-1 adapted more rapidly than WH86 in response to the low temperature treatment.
Chilling stress survival in Gr89-1 and WH86. Seedlings of WH86 and Gr89-1 were grown to three-leaf stage under regular growth conditions (see Materials and Methods) a and then moved to 4 °C (cold conditions) for 48 h and thereafter allowed to recover for 1 week b. c Chlorophyll fluorescence measurements. Three-leaf stage plants were used for the analysis. Plants were dark acclimated for 1 h and then moved to + 4 °C. Chlorophyll fluorescence measurements were
Transcriptome Profiling in Response to Chilling Stress in Seedlings
To study cold-induced changes in gene expression of the cold-tolerant genotype, the transcriptomes of Gr89-1 at six different cold treatment time points (0, 2, 6, 12, 24, and 48 h) were studied using the Illumina sequencing platform. The main features of these six libraries are summarized in Supplementary Table S3. For each library, 437,609 distinct tag sequences were generated (~ 12 million sequence tags in total). After removing the low-quality reads and trimming the adapter sequences, we obtained 221,840 distinct clean tag sequences (~ 11.7 million total clean sequence tags for each library) (Supplementary Table S3). Using Trinity for the transcriptome assembly, we unambiguously mapped 32.89–36.46% of the distinct clean tags to the UniGene database and 22.66–29.66% to the rice genome database; 15.21–21.83% of the distinct clean tags could not be mapped to the UniGene virtual tag database (Supplementary Table S3).
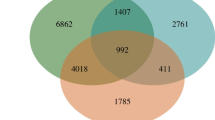
Genes presenting more than fourfold expression changes relative to the control level were identified as DEGs. In total, we identified 557, 1089, 1325, 1867, and 2342 DEGs at 2, 6, 12, 24, and 48 h, respectively, after chilling stress (Fig. 2b and Supplementary Table S4); 342 DEGs were significantly regulated at all five time points, and 16, 84, 118, 212, and 683 DEGs were present at 2, 6, 12, 24, and 48 h after chilling stress, respectively (Fig. 2a).
To explore the potential functions of the DEGs, functional annotation GO analysis was performed. All 2993 DEGs were divided into three categories (cellular components, molecular functions, and biological processes). The majority of the DEGs annotated as cellular components were categorized into cell part (GO: 0044464), organelle (GO: 0043226), and nuclear (GO: 0005634) subgroups. The majority of the DEGs annotated as molecular functions were categorized into catalytic activity (GO: 0003824), binding (GO: 0005488), and transcriptional regulatory activity (GO: 0030528) subgroups. The DEGs assigned to the catalytic activity subgroup mainly showed transferase, hydrolase, kinase, and oxidoreductase activities. The most common substances in the subgroups were carbohydrate, nucleosides, ions, tetrapyrroles, hemes, and proteins. The DEGs annotated as biological processes were mainly categorized into metabolic (GO: 0008152), cellular (GO: 0009987), and regulation of biological process (GO: 0050789) subgroups. The transport (GO: 0006810) and transcription (GO: 0006350) terms were also enriched within the biological processes category (Fig. 3). The phenylpropanoid biosynthesis (ko00940), plant hormone signal transduction (ko04075), protein processing in endoplasmic reticulum (ko04141), glutathione metabolism (ko00480), MAPK signaling pathway (ko04016), and starch and sucrose metabolism (ko00500) were significantly affected during low temperature (Fig. 4).
Chilling Stress Responsive Genes in Gr89-1 Seedlings
JA Metabolism and Signaling-Related Genes
In this study, 82 genes were involved in different plant hormone biosynthetic and signal transduction pathways during chilling stress, including jasmonic acid(JA), brassinosteroids (BRs), gibberellins (GAs), ABA, cytokinin (CK), ethylene (ET), salicylic acid (SA), and auxin (Figs. 5 and 6, Supplementary Table S5). These data indicated that a collaboration between phytohormone biosynthesis and signal transduction might be involved in the Gr89-1 seedlings during chilling tolerance. Thirteen DEGs were annotated as JA metabolism or signal transduction terms, including three LOXs (LOC_Os03g52860, LOC_Os04g37430, and LOC_Os08g39840) and three OPRs (LOC_Os06g11290, LOC_Os06g11260, and LOC_Os06g11210). Notably, almost all of the biosynthesis of JA genes was upregulated during cold acclimation (Fig. 5a and b, and Supplementary Table S5). Five JAZs (LOC_Os03g08310, LOC_Os03g08320, LOC_Os03g08330, LOC_Os10g25230, and LOC_Os10g25290) involved in the JA signaling pathway were upregulated during chilling stress in Gr89-1. The qRT-PCR results also showed that the LOXs, OPRs, and JAZs were strongly upregulated in rice seedlings (Fig. 5d and Supplementary Table S5). The transcriptional data suggested that JA accumulated under cold stress.
Overview of putative JA biosynthesis pathways involved in rice and expression profiles of genes involved in this pathway. a Expression of genes involved in JA metabolism and signaling under low temperature conditions. b Biosynthetic pathway of JA in rice. Genes that are up- or downregulated by > 2-fold after stress are indicated in red and blue, respectively. c JA contents in Gr89-1 and WH86 leaves determined by HPLC-MS. d Relative expression of genes involved in JA metabolism pathways. TPM: number of transcripts per million tags. Error bars indicate the SD for three independent replicates. LOX, lipoxygenase; OPR, 12-OPDA, (15Z)-12-Oxophytodienoic acid; OPC-8, 3-oxo-2-(2-pentenyl)-cyclopentane-1-octanoic acid; JMT, S-adenosyl-L-methionine:jasmonic acid carboxyl methyltransferase; 12-oxophytodienoate reductase; MeJA, methyl-JA; TIFY, ZIM domain-containing protein
DEG enrichment of the plant hormone signal transduction pathway by KEGG annotation. The key regulatory components in multiple hormone response pathways are presented as their names (red, upregulated; green, downregulated). Gene IDs and fold changes in transcript abundance are indicated in Supplementary Table S5
Therefore, we measured the content of endogenous JA in Gr89-1 and WH86 seedlings by HPLC-MS in this study. It was worth noting that the content of JA was higher in Gr89-1 compared with WH86 under cold stress. Moreover, Gr89-1 showed a more sharply increased JA concentration after 24 h of cold treatment compared with WH86 (Fig. 5c). These results show that low temperature stress may influence the homeostasis of endogenous JA in rice seedlings, and the higher concentration of JA was detected in Gr89-1. These results show that activation of the JA biosynthetic pathway plays a key role in the cold tolerance of Gr89-1. The HPLC-MS and qRT-PCR results confirmed the reliability of the digital expression data.
Other Plant Hormones in Chilling Stress
In addition to JA, 69 genes were involved in different plant hormone biosynthetic and signal transduction pathways, including brassinosteroids (BRs), gibberellins (GAs), ABA, cytokinin (CK), ethylene (ET), salicylic acid (SA), and auxin (Supplementary Table S5). The DEGs for ET biosynthesis and signal transduction were upregulated. The expression of phenylalanine ammonia-lyase (PAL; LOC_Os02g41650 and LOC_Os04g43760), encoding an SA synthetic enzyme, was induced and the cytoplasmic binding protein non-expressor of pathogenesis-related genes (NPR1, LOC_Os03g46440) was upregulated. The PR-1 gene (LOC_Os10g11500), which encodes downstream proteins of the SA signaling cascade, was also upregulated at 2 h. BR, GA, ABA, CK, and auxin also participated in the chilling stress (Fig. 6 and Supplementary Table S5).
Protein Kinases
In seedlings, the DEGs involved in calcium signaling included calmodulin (CML), calcineurin B-like protein-interacting protein kinases, calcium-dependent protein kinases (CDPKs), and mitogen-activated kinases (Fig. 7 and Supplementary Table S6). It is worth emphasizing that the expression of all of CML, MAPK, MAPKK, and MAPKKK genes was upregulated at all five cold exposure (4 °C) time points, particularly LOC_Os02g58520, LOC_Os12g01400, LOC_Os01g50400, and LOC_Os05g46760, which displayed an at least 5.09-fold increase at all time points (Fig. 7 and Supplementary Table S6).
Transcription Factor-Related Genes
In total, 243 TF genes in Gr89-1 were identified during at least one chilling stress time point (Fig. 8 and Supplementary Table S7). We found 35 differentially regulated AP2/EREBP genes after 2 h of cold exposure, of which 25 were upregulated strongly. In particular, DREB1A (LOC_Os09g35030), DREB1B (LOC_Os09g35010), DREB1C (LOC_Os06g03670.1), DREB1F (LOC_Os01g73770), DREB1G (LOC_Os02g45450), DREB1H (LOC_Os09g35020), OsEATB (LOC_Os09g28440), and EREBP2 (LOC_Os01g64790) were highly upregulated in Gr89-1. A total of 109 TF genes from the MYB, NAC, bHLH, WRKY, C2H2, HB, and bZIP families were differentially expressed in seedlings, mostly after 2 h of cold exposure, whereas downregulation of all GNAT and mTERF family genes occurred, where the roles of these genes in chilling stress are unclear (Fig. 8 and Supplementary Table S7).
Osmotic Adjustment-Related Genes
Changes in osmoprotectants represent a typical physiological and biochemical response to stress. Genes involved in soluble sugar metabolism exhibited differential expression upon exposure to low temperature (Fig. 9 and Supplementary Table S8). Genes encoding β-amylase (LOC_Os10g41550), sucrose-phosphate synthase (LOC_Os02g09170), and sucrose synthase (LOC_Os04g24430) (5.7-fold after 2 h at 4 °C) were specifically upregulated in Gr89-1 under the chilling stress treatment. Several genes related to trehalose and stachyose biosynthesis were upregulated (Fig. 9 and Supplementary Table S8). Among the genes related to trehalose biosynthesis, two trehalose-6-phosphate synthases (TPS; LOC_Os08g34580 and LOC_Os08g31980), and one trehalose-6-phosphate phosphatase (LOC_Os08g31630) were upregulated in Gr89-1. Additionally, one stachyose synthase (LOC_Os01g07530) involved in stachyose biosynthesis was upregulated under chilling stress in Gr89-1 seedlings (Fig. 9 and Supplementary Table S8). Therefore, we measured whether the total soluble sugar, sucrose, and trehalose were also increased during the cold acclimation process (Fig. 9b). The quantitative determination of oligosaccharides analysis showed that the total soluble sugar contents increased sharply after 6 h of chilling stress and was maintained at a high level from 6 to 48 h compared with control seedlings (Fig. 2b). The sucrose level increased gradually during cold treatment and increased accumulation of trehalose after 12 h of chilling stress (Fig. 9b). These data indicate that the total soluble sugar, sucrose, and trehalose play vital roles in the cold resistance of Gr89-1.
Starch and sucrose metabolisms in Gr89-1 seedlings subject to chilling stresses. a Expression of genes involved in starch and sucrose metabolism under low temperature conditions. b The total soluble sugar, sucrose, and trehalose contents in the seedling of Gr89-1 under normal temperature and cold treatments
Cell Wall-Related Genes
The genes related to the cell wall showing altered expression patterns were divided into precursor generation, polysaccharide synthesis, and assembly subgroups (Fig. 10 and Supplementary Table S9). During precursor synthesis, the enzyme UDP-glucuronate 4-epimerase, which catalyzes the transformation of UDP-α-glucose to UDP-α-galactose, and LOC_Os01g62020, LOC_Os02g54890, and LOC_Os08g41440, were upregulated after 2 h of chilling stress. Among the polysaccharide synthesis-related genes, two genes for hemicellulose synthase (CSLC-9: LOC_Os03g56060; GT47: LOC_Os03g20850) and four genes encoding pectin biosynthetic (GT8: LOC_Os02g50600, LOC_Os03g21250, LOC_Os03g47530, and LOC_Os08g23780) genes were activated in seedlings. Notably, the expression of the transcript for LOC_Os03g47530 was over sevenfold higher at the beginning of exposure to 4 °C. We observed that several genes involved in cell wall assembly were activated and/or repressed in seedlings under cold stress. Five genes in the expansin (EXP) family were downregulated under cold stress. Among the nine DEGs in the xyloglucan endotransglucosylases/hydrolase (XTH) family, most were upregulated in rice seedlings under cold stress. The 19 glycosyl hydrolase (GH) family genes identified were mainly from GH17; LOC_Os01g71860 and LOC_Os06g34020 showed an at least 4.09-fold increase after 2 h of cold exposure. The esterase LOC_Os03g18860 was upregulated 9.64-fold after 48 h at 4 °C. Similarly, LOC_Os10g05820, which belongs to the proline-rich protein (PRP) gene family, also showed strongly increased expression after 6 h of cold treatment (Fig. 10 and Supplementary Table S9).
Discussion
Rice is one of the most important food crops in China. Low temperature is a major factor affecting rice geographical distribution growth, development, quality, and productivity. It is significant to study the molecular mechanism of low-temperature tolerance of rice and cultivate high-yield and high-quality cold-resistant varieties to ensure food security in China. In this study, Gr89-1, which tolerates low temperatures and has overwintering ability, was used to investigate the molecular mechanism of rice response to chilling stress at seedling stage by RNA-seq technology. After comprehensive analysis of DEGs, we inferred that JA biosynthetic and signal transduction, sucrose, stachyose, and trehalose synthesis, and cell structure and synthesis were particularly influenced in Gr89-1 seedlings under cold stress.
Phytohormones, such as ABA, GA, SA, auxin, and ET, are positive or negative regulators of the chilling stress response (Miura and Furumoto 2013). In this study, collaboration between phytohormone biosynthesis and signal transduction might be involved in the Gr89-1 seedlings during chilling tolerance (Figs. 5 and 6, Supplementary Table S5). The phytohormone JA regulates plant responses to biotic and abiotic stressors (Wasternack and Hause 2013). Exogenous application of MeJA increases the chilling tolerance in Arabidopsis, banana, pomegranate fruit, and wheat (Hu et al. 2013; Qi et al. 2006; Zhao et al. 2013; Zolfagharinasab and Hadian 2007). Exposure to cold rapidly accumulates endogenous JA content by inducing JA biosynthesis genes such as LOX1, AOS1, AOC1, AOC2, and JAR1 in Arabidopsis (Goulas et al. 2006; Hu et al. 2013) and OsAOS, OsOPR1, OsAOC, and OsLOX2 in rice (Du et al. 2013). Compared with wild-type plants, Arabidopsis mutants (lox2, aos, jar1, and coi1) deficient in JA biosynthesis or signaling are more sensitive to freezing stress (Hu et al. 2013). In C. japonica, nine JA biosynthesis genes are induced during cold acclimation (Li et al. 2016). Similarly, the expression profiles of our six JA-biosynthetic genes (three LOXs: LOC_Os03g52860, LOC_Os04g37430, and LOC_Os08g39840, and three OPRs: LOC_Os06g11290, LOC_Os06g11260, and LOC_Os06g11210) indicated significant upregulation during cold exposure in this study, and free JA content was upregulated in 4 °C-treated seedlings of Gr89-1 after 24 h (Fig. 5c). It has also reported that the Ca2+ and MAPK cascades are also involved in the regulation of JA biosynthesis (Wasternack and Hause 2013). CDPKs act as positive regulators of plant Ca2+-regulated cold signaling (Saijo et al. 2010). The induction of CML genes by chilling stress has been described in wild rice (Zhang et al. 2017a, b). In our study, 12 CDPK and 8 CML genes were differentially expressed under severe chilling stress, and most of these genes were activated (Fig. 7 and Supplementary Table S6). The mitogen-activated protein kinase (MAPK) cascade is implicated in the regulation of cold signaling and freezing tolerance in plants (Teige et al. 2004). In this study, 12 MKKK, 2 MKK, and 2 MAPK genes showed increased expression during cold exposure (Fig. 7 and Supplementary Table S6), particularly LOC_Os01g50400 (6.27-fold increase after 2 h at 4 °C). Therefore, we speculated that the Ca2+ and MAPK cascade pathway may be involved in chilling tolerance in Gr89-1 seedlings.
Under cold stress, increased JA content triggers COI1-mediated degradation of JAZs, and this releases ICEs from repression (Hu et al. 2017). JA acts as an upstream signal in the ICE-CBF/DREB1 pathway, which positively regulates cold tolerance in Arabidopsis and tomato (Hu et al. 2013; Wang et al. 2016). Several TF family members, such as AP2/EREBP, NAC, bHLH, R2R3-MYB, and WRKY, are recruited in JA signaling (Goulas et al. 2006). According to our data, 35 AP2/EREBP genes were differentially regulated at low temperature, of which 25 were upregulated in seedlings (Fig. 8 and Supplementary Table S7). OsDREB1A (LOC_Os09g35030), OsDREB1B (LOC_Os09g35010), and OsDREB1C (LOC_Os06g03670) were highly upregulated in Gr89-1, consistent with previous reports that CBF/DREB1 has a central role in chilling tolerance in plants (Chinnusamy et al. 2007; Dubouzet et al. 2003). In addition, most DEGs encoding WRKY, MYB, NAC, and C2H2 TF genes were upregulated in our study, indicating that induction of the AP2/ERF, WRKY, MYB, NAC, and C2H2 families of genes was required for chilling stress tolerance of Gr89-1. Therefore, we inferred that the significant JA accumulation may play a positive role in Gr89-1 adaptation to low temperatures.
A large number of osmoprotectants are synthesized downstream of the transcriptional regulatory network (Krasensk et al. 2012). Overexpression of CBF3 increases levels of proline and total soluble sugars in non-acclimated and cold-acclimated Arabidopsis (Achard et al. 2008; Cook et al. 2004; Gilmour et al. 2004, 2000). As osmoprotectants and nutrients, soluble sugars are activated to protect the plasma membrane against damage caused by chilling stress (Tarkowski and Van den Ende 2015). In this study, the total soluble sugar contents increased sharply after 6 h of chilling stress and were maintained at a high level from 6 to 48 h compared with control seedlings (Fig. 9 and Supplementary Table S8). Overexpression of OsTPP1 and OsTPS1 increases levels of trehalose in rice, and activation of trehalose biosynthesis increases plant cold resistance (Ge et al. 2008; Habibur Rahman Pramanik and Imai 2005; Li et al. 2011; Miranda et al. 2007). Higher concentration of trehalose and significantly upregulated expression of the key enzymes involved in the biosynthesis of trehalose was detected in Gr89-1 seedlings (Fig. 9 and Supplementary Table S8). These data indicated that these soluble sugars may have major impacts on osmotic adjustment during cold stress in Gr89-1 seedlings. OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination by OsHOS1, and OsbHLH002 activates OsTPP1 and enhances the cold tolerance of rice (Zhang et al. 2017a, b). Therefore, we speculated that the MAPK cascade pathway involved in chilling tolerance in Gr89-1 seedlings by increasing soluble sugars.
Undoubtedly, cold stress has a profound effect on cell wall polysaccharide composition and cell wall-modifying enzyme activities (Miura and Furumoto 2013). Pectin is a major polysaccharide of primary cell walls and is involved in wall plasticity and cellular adhesion. Pectin content was shown to increase in response to low temperatures in oilseed rape, leaves, pea epicotyls, and bromeliads (Carvalho et al. 2013; Kubacka-Zębalska and Kacperska 1999; Solecka et al. 2008; Weiser et al. 1990). In this study, four pectin biosynthesis-related enzymes were upregulated during cold exposure (Fig. 10 and Supplementary Table S9), particularly LOC_Os03g47530 (7.86-fold after 2 h at 4 °C). Cell wall-modifying enzymes, such as pectin methylesterase (CE8), polygalacturonase (GH28), pectin/pectate lyase-like (PL1), and pectin acetylesterase (CE13), are involved in cell wall plasticity/rheology (Sénéchal et al. 2014). All of these enzymes were differentially expressed during cold exposure (Fig. 6 and Supplementary Table S9). EXPs (Cosgrove 2005; Marowa et al. 2016), GHs (Buchanan et al. 2012; Glass et al. 2015; Wei et al. 2015), and XTHs (Nishitani and Vissenberg 2006; Rose et al. 2002) participate in the cell wall loosening process (Li et al. 2003; Minic and Jouanin 2006). EXP genes are activated in rice and Arabidopsis, while EXPs and XTHs are repressed in sweet potato (Imin et al. 2004; Noh et al. 2009; Yamauchi et al. 2004). Overexpression of AtXTH21 improves freezing tolerance in transgenic Arabidopsis plants (Shi et al. 2014). In the present study, the expression of EXP genes was downregulated, while that of XTHs and GHs (GH9, GH17, GH18, and GH28) was upregulated in seedlings (Fig. 5 and Supplementary Table S9). Cold stress also induces changes in plant cell wall proteins, such as hydroxyproline-rich glycoproteins (HRGPs). HRGPs are classified into three subfamilies: AGPs, EXTs, and PRPs (Showalter et al. 2010). Overexpression of GhAGP31 improves cold tolerance in transgenic Arabidopsis seedlings (Gong et al. 2012). In our study, nine AGP, seven EXT, and eight PRP genes were differentially expressed in seedlings exposed to cold stress, indicating that the expression of cell wall-related genes enhances the low temperature resistance in Gr89-1 seedlings.
By analyzing transcriptional profile, we found that increase in endogenous JA content and osmoprotectants and change cell wall composition in seedlings under chilling stress could enhance the cold tolerance of Gr89-1. These results may offer clues for improvement mechanisms of response to low temperature and cultivation regulation at rice seedling stage.
Change history
13 November 2021
A Correction to this paper has been published: https://doi.org/10.1007/s11105-021-01322-6
References
Achard P, Gong F, Cheminant S, Alioua M, Hedden P, Genschik P (2008) The cold-inducible CBF1 factor–dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20:2117–2129
Baruah AR, Ishigo-Oka N, Adachi M, Oguma Y, Tokizono Y, Onishi K, Sano Y (2009) Cold tolerance at the early growth stage in wild and cultivated rice. Euphytica 165:459–470
Buchanan M, Burton RA, Dhugga KS, Rafalski AJ, Tingey SV, Shirley NJ, Fincher GB (2012) Endo-(1,4)-β-Glucanase gene families in the grasses: temporal and spatial co-transcription of orthologous genes. BMC Plant Biol 12:235
Buti M, Pasquariello M, Ronga D, Milc JA, Pecchioni N, Ho VT, Pucciariello C, Perata P, Francia E (2018) Transcriptome profiling of short-term response to chilling stress in tolerant and sensitive Oryza sativa ssp. Japonica seedlings Funct Integr Genomics 18:627–644
Carvalho CP, Hayashi AH, Braga MR, Nievola CC (2013) Biochemical and anatomical responses related to the in vitro survival of the tropical bromeliad Nidularium minutum to low temperatures. Plant Physiol Biochem 71:144–154
Chawade A, Lindlöf A, Olsson B, Olsson O (2013) Global expression profiling of low temperature induced genes in the chilling tolerant japonica rice Jumli Marshi. PLoS One 8:e81729
Chinnusamy V, Zhu J, Zhu JK (2007) Cold stress regulation of gene expression in plants. Trends Plant Sci 12:444–451
Cook D, Fowler S, Fiehn O, Thomashow MF (2004) A prominent role for the CBF cold response pathway in configuring the low-temperature metabolome of Arabidopsis. Proc Natl Acad Sci U S A 101:15243–15248
Cosgrove DJ (2005) Growth of the plant cell wall. Nat Rev Mol Cell Biol 6:850–861
Deng XS, Gan L, Liu Y, Luo AC, Jin L, Chen J, Tang RY, Lei LX, Tang JH, Zhang JN, Zhao ZW (2018) Locating QTLs controlling overwintering seedling rate in perennial glutinous rice 89–1 (Oryza sativa L.). Genes & Genomics 40:1351–1361
Du H, Liu HB, Xiong LZ (2013) Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front Plant Sci 4:397
Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J 33:751–763
Ge LF, Chao DY, Shi M, Zhu MZ, Gao JP, Lin HX (2008) Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 228(1):191–201
Gilmour SJ, Fowler SG, Thomashow MF (2004) Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol Biol 54:767–781
Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF (2000) Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol 124:1854–1865
Glass M, Barkwill S, Unda F, Mansfield SD (2015) Endo-β-1,4-glucanases impact plant cell wall development by influencing cellulose crystallization. J Integr Plant Biol 57:396–410
Gong SY, Huang GQ, Sun X, Li P, Zhao LL, Zhang DJ, Li XB (2012) GhAGP31, a cotton non-classical arabinogalactan protein, is involved in response to cold stress during early seedling development. Plant Biol 14:447–457
Goulas E, Schubert M, Kieselbach T, Kleczkowski LA, Gardeström P, Schröder W, Hurry V (2006) The chloroplast lumen and stromal proteomes of Arabidopsis thaliana show differential sensitivity to short- and long-term exposure to low temperature. Plant J 47:720–734
Habibur Rahman Pramanik M, Imai R (2005) Functional identification of a trehalose 6-phosphate phosphatase gene that is involved in transient induction of trehalose biosynthesis during chilling stress in rice. Plant Mol Biol 58:751–762
He GC, Shu LH, Zhou YQ, Liao LJ (1996) The overwintering ability of Dongxiang wild rice (Oryza rufipogon) at Wuhan. Wuhan University Journal 42:252–254 (in Chinese)
Hu YR, Jiang LQ, Wang F, Yu DQ (2013) Jasmonate regulates the inducer of cbf expression-C-repeat binding factor/DRE binding factor 1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25:2907–2924
Hu YR, Jiang YJ, Han X, Wang HP, Pan JJ, Yu DQ (2017) Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J Exp Bot 68:1361–1369
Imin N, Kerim T, Rolfe BG, Weinman JJ (2004) Effect of early cold stress on the maturation of rice anthers. Proteomics 4:1873–1882
Jeon J, Kim J (2013) Cold stress signaling networks in Arabidopsis. J Plant Biol 56:69–76
Kim J (2007) Perception, transduction, and networks in cold signaling. J Plant Biol 50:139–147
Krasensky J, Jonak C (2012) Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J Exp Bot 63:1593–1608
Kubacka-Zębalska M, Kacperska A (1999) Low temperature-induced modifications of cell wall content and polysaccharide composition in leaves of winter oilseed rape (Brassica napus L. var. oleifera L.). Plant Sci 148:59–67
Li HW, Zang BS, Deng XW, Wang XP (2011) Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 234(5):1007–1018
Li QY, Lei S, Du KB, Li LZ, Pang XF, Wang ZC, Wei M, Fu S, Hu LM, Xu L (2016) RNA-seq based transcriptomic analysis uncovers α-linolenic acid and jasmonic acid biosynthesis pathways respond to cold acclimation in Camellia japonica. Sci Rep 6:36463
Li Y, Jones L, McQueen-Mason S (2003) Expansins and cell growth. Curr Opin Plant Biol 6:603–610
Liang YS, Zheng J, Yan C, Li XX, Liu SF, Zhou JJ, Qin XJ, Nan WB, Yang YQ, Zhang HM (2017) Locating QTLs controlling overwintering trait in Chinese perennial Dongxiang wild rice. Mol Genet Genomics 293:81–93
Lissarre M, Ohta M, Sato A, Miura K (2010) Cold-responsive gene regulation during cold acclimation in plants. Plant Signal Behav 5:948–952
Liu HB, Li DQ, Du H, Yuan M (2018) Quantification analysis of IAA, JA. ABA and SA in rice Bio- 101:e1010156. https://doi.org/10.21769/BioProtoc.1010156(inChinese)
Mao DH, Yu L, Chen DZ, Li LY, Zhu YX, Xiao YQ, Zhang DC, Chen CY (2015) Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor Appl Genet 128:1359–1371
Marowa P, Ding A, Kong Y (2016) Expansins: roles in plant growth and potential applications in crop improvement. Plant Cell Rep 35:949–965
Minic Z, Jouanin L (2006) Plant glycoside hydrolases involved in cell wall polysaccharide degradation. Plant Physiol Biochem 44:435–449
Miranda JA, Avonce N, Suárez R, Thevelein JM, Dijck PV, Iturriaga G (2007) A bifunctional TPS-TPP enzyme from yeast confers tolerance to multiple and extreme abiotic-stress conditions in transgenic Arabidopsis. Planta 226:1411–1421
Miura K, Furumoto T (2013) Cold signaling and cold response in plants. Int J Mol Sci 14:5312–5337
Nishitani K, Vissenberg K (2006) Roles of the XTH protein family in the expanding cell. In: Verbelen JP. and Vissenberg K. (eds) The Expanding Cell. Plant Cell Monographs, vol 6. Springer, Berlin, Heidelberg, pp 89–116
Noh SA, Park SH, Huh GH, Paek KH, Shin JS, Bae JM (2009) Growth retardation and differential regulation of expansin genes in chilling-stressed sweet potato. Plant Biotechnol Rep 3:75–85
Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L (2007) The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res, 35(Database issue):883–887
Peleg Z, Blumwald E (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr Opin Plant Biol 14:290–295
Pradhan SK, Pandit E, Nayak DK, Behera L, Mohapatra T (2019) Genes, pathways and transcription factors involved in seedling stage chilling stress tolerance in indica rice through RNA-Seq analysis. BMC Plant Biol 19:352
Qi FG, Li JM, Duan LS, Li ZH (2006) Inductions of coronatine and MeJA to low-temperature resistance of wheat seedlings. Acta Botanica Boreali-Occidentalia Sinica 26:1776–1780 (in Chinese)
Qian X, Truong TT, Barrero JM, Jacobsen JV, Hocart CH, Frank G (2016) A role for jasmonates in the release of dormancy by cold stratification in wheat. J Exp Bot 67:3497–3508
Romualdi C, Bortoluzzi S, D’Alessi F, Danieli GA (2003) IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics 12:159–162
Rose JKC, Janet B, Fry SC, Kazuhiko N (2002) The XTH family of enzymes involved in xyloglucan endotransglucosylation and endohydrolysis: current perspectives and a new unifying nomenclature. Plant Cell Physiol 43:1421–1435
Saijo Y, Hata S, Kyozuka J, Shimamoto K, Izui K (2010) Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J 23:319–327
Sénéchal F, Wattier C, Rustérucci C, Pelloux J (2014) Homogalacturonan-modifying enzymes: structure, expression, and roles in plants. J Exp Bot 65:5125–5160
Shi HT, Ye TT, Zhong B, Liu X, Jin R, Chan ZL (2014) AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytol 9:554–567
Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6:410–417
Showalter AM, Brian K, Jens L, Dazhang G, Welch LR (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol 153:485–513
Solecka D, Zebrowski J, Kacperska A (2008) Are pectins involved in cold acclimation and de-acclimation of winter oil-seed rape plants? Ann Bot 101:521–530
Tarkowski ŁP, Van den Ende W (2015) Cold tolerance triggered by soluble sugars: a multifaceted countermeasure. Front Plant Sci 6
Teige M, Scheikl E, Eulgem T, Dóczi R, Ichimura K, Shinozaki K, Dangl JL, Hirt H (2004) The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol Cell 15:141–152
Thomashow MF (1999) Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu Rev Plant Physiol Plant Mol Biol 50:571–599
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Trapnell C, Roberts A, Gof L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Wang CP, Lei KR, Li ZG, Lin Q, Wu H (2012) Effects of chilling stress on chlorophyll fluorescence characteristics of seedling leaves with different leaf ages of Oryza sativa. J Plant Res Environ 21:38–43 (in Chinese)
Wang F, Guo ZX, Li HZ, Wang MM, Onac E, Zhou J, Xia XJ, Shi K, Yu JQ, Zhou YH (2016) Phytochrome A and B function antagonistically to regulate cold tolerance via abscisic acid-dependent jasmonate signaling. Plant Physiol 170:459–471
Wasternack C, Hause B (2013) Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann Bot 111:1021–1058
Wei H, Brunecky R, S. Donohoe B, Ding SY, N. Ciesielski P, Yang SH, P. Tucker M, E. Himmel M (2015) Identifying the ionically bound cell wall and intracellular glycoside hydrolases in late growth stage Arabidopsis stems: implications for the genetic engineering of bioenergy crops. Front Plant Sci 6
Weiser RL, Wallner SJ, Waddell JW (1990) Cell wall and extensin mRNA changes during cold acclimation of pea seedlings. Plant Physiol 93:1021–1026
Yamauchi Y, Ogawa M, Kuwahara A, Hanada A, Kamiya Y, Yamaguchi S (2004) Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16:367–378
Yoshida S, Forno DA, Cock JH, Gomez KA (1972) Laboratory manual for physiological studies of rice. IRRI, Manila, Philippines, pp 67–68
Zhang F, Huang LY, Wang WS, Zhao XQ, Zhu LH, Fu BY, Li ZK (2012) Genome-wide gene expression profiling of introgressed indica rice alleles associated with seedling cold tolerance improvement in a japonica rice background. BMC Genomics 13:461
Zhang T, Huang LY, Wang YX, Wang WS, Zhao XQ, Zhang SL, Zhang J, Hu FY, Fu BY, Li ZK (2017) Differential transcriptome profiling of chilling stress response between shoots and rhizomes of Oryza longistaminata using RNA sequencing. PLoS One 12:e0188625
Zhang T, Zhao XQ, Wang WS, Pan YJ, Huang LY, LiuZ Y, Zhu LH, Yang DC, Fu BY, XY (2012) Comparative transcriptome profiling of chilling stress responsiveness in two contrasting rice genotypes. PLoS One 7:e43274
Zhang ZY, Li JH, Li F, Liu HH, Yang WS, Chong K, Xu YY (2017) OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell 43:731–743
Zhao ML, Wang JN, Shan W, Fan JG, Kuang JF, Wu KQ, Li XP, Chen WX, He FY, Chen JY, Lu WJ (2013) Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ 36:30–51
Zolfagharinasab R, Hadian J (2007) Influence of methyl jasmonate on inducing chilling tolerance in pomegranate fruits (Malas Save). Pak J Biol Sci 10:612–616
Funding
The research was financially supported by the performance incentive guide special project of Chongqing Academy of Agricultural Sciences (cstc2018jxjl80026).
Author information
Authors and Affiliations
Contributions
KL conceived and designed the experiments. XP and HW carried out the experiments. WZ, XJ, LG, and WB analyzed the data. XP and KL wrote and revised the paper. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Key Message
• Glutinous rice 89-1 (Gr89-1) improves its response to chilling stress by increasing the production of osmoprotectants (soluble sugar, sucrose, and trehalose), modifying its cell wall structure and composition, and altering the JA biosynthetic and signal transduction pathways.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Pan, X., Wu, H., Hu, M. et al. Global Analysis of Gene Expression Profiles in Glutinous Rice 89-1 (Oryza sativa L.) Seedlings Exposed to Chilling Stress. Plant Mol Biol Rep 39, 626–639 (2021). https://doi.org/10.1007/s11105-020-01278-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01278-z