Abstract
Larix olgensis is one of the main coniferous tree species in northeastern China and has excellent timber properties and strong tolerance to stress. Thirteen HD-Zip family genes with a complete CDS region were identified on the basis of cambium transcriptome data from Larix olgensis. All 13 genes were analyzed via bioinformatics by their conserved domain protein sequence and amino acid composition, including their physicochemical properties and protein structure. The spatiotemporal expression and abiotic stress responses of these genes were analyzed by real-time quantitative PCR. The results showed that the 13 HD-Zip genes of Larix olgensis were expressed in the roots, stems, and leaves at different stages. The expression of three of these genes (LoHDZ2, LoHDZ11, LoHDZ13) was highest in nonlignified roots, indicating that they might be related to the secondary growth of Larix olgensis; in addition, three genes (LoHDZ5, LoHDZ9, LoHDZ10) were highly expressed in partially and completely lignified stems and leaves. These 13 genes were expressed specifically under drought stress. The expression of two of them (LoHDZ1, LoHDZ5) was obviously upregulated, and the expression of 6 genes (LoHDZ2, LoHDZ3, LoHDZ4, LoHDZ8, LoHDZ10, LoHDZ13) was significantly downregulated. The expression trends indicate that these genes could be involved in drought stress. The expression of all 13 genes was downregulated when the plants were treated with 0.2 M NaCl for 96 h, indicating that these genes are inhibited by salt stress. Overall, the results have significant implications for the study of the gene function of members of the LoHD-Zip transcription factor family.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Larix olgensis is a deciduous tree species in the genus Larix and is the main timber and afforestation species in Northeast China (Yu and Bao 2010). High salinity, drought, and low temperature constitute the main abiotic factors affecting the growth of larch and other tree species in Northeast China (Jinhua et al. 2003). Studies on the molecular mechanisms involving these genes can unravel the mystery of plant growth under adverse conditions, and genetic engineering can be used to improve the ability of plants to resist various abiotic stresses. The present research on gene function mainly focuses on functional genes and transcription factors. Transcription factors, which are also known as trans-acting factors, are DNA-binding proteins that can specifically interact with cis-acting elements within the promoter region of eukaryotic genes and activate or inhibit transcription via interactions between them and with other related proteins (Liu et al. 2008). Studies have shown that many transcription factors are involved in the regulation of plant growth and abiotic stress, such as HD-Zip WRKY, AP2/ERF, and MYB (Schena and Davis 1992).
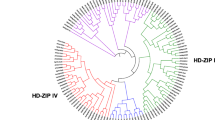
Homeodomain-leucine zippers (HD-Zip) are a kind of plant-specific transcription factor. HD-Zip belongs to the homeobox protein family and has a highly conserved homeodomain (HD). Consisting of 61 amino acids (Schena and Davis 1992), this domain can be folded into a typical spiral-ring-helix-turn-helix structure that can specifically bind to DNA (Bharathan et al. 1997). Its end is closely linked to the leucine zipper (LZ) domain, and the leucine zipper domain mainly mediates the formation of protein dimers (Jain et al. 2008). This indicates that HD-Zip genes play an important role in certain developmental processes of higher plants (Ariel et al. 2007). Based on sequence conservation, gene structure and physiological functions, HD-Zip transcription factors can be divided into four subclasses: HD-ZipI, HD-ZipII, HD-ZipIII, and HD-ZipIV (Mukherjee et al. 2009) (Fig. 1). Due to the differences in gene composition and the structure of their encoded proteins in different subfamilies, they are involved in different plant development processes and regulate different metabolic processes. Current research on the HD-Zip transcription factor family in Arabidopsis, rice, apple, alfalfa, and other plant species has revealed several conclusions: HD-ZipI proteins may be involved in light signal transduction (Henriksson et al. 2005), regulation of leaf and seed development (Henriksson et al. 2005; Kim et al. 2007), and responses to stress (Harris et al. 2016; Zhao et al. 2014a, b); the conserved domain unique to the HD-Zip class II subfamily comprises CPSCE (Ciarbelli et al. 2008) and plays an important role in mediating responses to light quality changes (Franklin et al. 2003), shade (Carabelli et al. 2013), and abiotic stress (Park et al. 2013); the HD-ZipIII subfamily genes have a complex structure that allows them to inhibit the transcription of genes mainly involved in plant embryo development (Prigge et al. 2005), meristem formation (Zhu et al. 2013), and vascular development (Robischon et al. 2011); and the HD-Zip class IV subfamily genes play an important role in plant epidermal cell development (Ingram et al. 2000), material accumulation and transport (Ito et al. 2002), and resistance to both biotic stress and abiotic stress (Nakamura et al. 2006).
In this study, BLAST alignments were performed via the NCBI website (https://blast.ncbi.nlm.nih.gov) by analyzing the results of the existing transcriptome data from the Larix olgensis cambium. All 13 HD-Zip transcription factor family genes (named LoHDZ1~LoHDZ13) with complete CDS regions were obtained. To understand the function of the members of the HD-Zip family of Larix olgensis, bioinformatic analysis and gene expression analysis were performed on these 13 HD-Zip genes. Because there are few studies on the larch HD-Zip transcription factor family, this study provides a partial reference for future studies on the functions of larch HD-Zip transcription factor family genes.
Materials and Methods
Materials
Seeds of Larix olgensis used in this experiment were obtained from Jixi, Heilongjiang Province. Full, lustrous Larix olgensis seeds were selected and soaked in deionized water for 4–5 days, which was replaced 3–4 times during this period. The plants were then planted in a soil to vermiculite to perlite (5:3:2 v/v/v) nutrient soil matrix (the substrates used in this experiment were nutrient-enriched components arranged at that ratio). Using plastic wrap to maintain sufficient moisture, the plant growth conditions consisted of a 16-h light/8-h dark photoperiod, a humidity of 75%, and a constant temperature of 22 °C.
After 25–30 days, when the needles of larch were fully extended, the seedlings exhibiting good growth were selected and transplanted to a seedling basin for further cultivation in the nutrient-enriched soil matrix. Seedlings grown for 2 months were treated with 7% (w/v), 14%, 21%, and 28% PEG6000 for 0 h (control), 24 h, 48 h, 72 h, and 96 h, at which times samples were collected (Lian et al. 2019). After processing, extracted RNA of complete seedlings for reverse transcription; other larch seedlings were sprayed with 0.2 M NaCl solution (Ma et al. 2012), after which samples were collected after 0 h (control), 12 h, 24 h, 48 h, and 96 h. After treatment, extracted RNA of complete seedlings for reverse transcription and real-time PCR was performed.
The roots, stems, and leaves (organ parts were taken from the same plant during the same period) of Larix olgensis plants that had grown for 3 months (no lignification period), 6 months (partial lignification period), and 9 months (complete lignification period) were collected. The organs were collected from the same plant as from which material was collected for the expression analysis of the genes in different tissues at different periods.
Methods
Bioinformatic Analysis
The study was based on the existing transcriptome data of Larix olgensis and compared 13 HD-Zip family genes with complete CDS regions using the NCBI website. Thirteen HD-Zip transcription factor family genes were classified according to their conserved domain characteristics by using BioEdit bioanalytical software. Thirteen genes were translated into their corresponding amino acid sequences by using BIOXM software. ExPASy online software was then used to analyze the physical and chemical properties of the LoHD-Zip protein and the molecular weight and isoelectric point of the 13 HD-Zip genes; moreover, the physicochemical properties, such as the fat coefficient and stability coefficient (Zhang et al. 2017), were obtained. The secondary structure of the protein was analyzed by SOPMA online software. And their subcellular localization was predicted by SUBA4 online software MEGA 5.0 which was used to construct an evolutionary tree of the 13 LoHD-Zip and 50 Arabidopsis HD-Zip family genes. The bioinformatics website is shown in Table 1.
Real-Time PCR
Fluorescence quantitative PCR was used to analyze the spatiotemporal expression pattern of the genes in larch and their patterns of involvement in the abiotic stress response.
In this study, the RNA of Larix olgensis was extracted by using a PureLink™ Plant RNA Reagent Kit, and the extraction procedure was carried out according to the manufacturer’s instructions; the RNA of Larix olgensis was then reverse transcribed using a kit (ReverseScriptRT reagent Kit, TaKaRa) to obtain cDNA. Primer Premier 5 software (Xin-Yu and Yan-Ning 2004) was used to design the primers (Table 2), and the fluorescence quantification reagent was TaKaRa’s SYBR Premix Ex TaqII (Tli RnaseH Plus) Kit. The test procedure was carried out according to the instructions of the kit (Table 3), and an ABI 7500 real-time PCR instrument (made by American ABI company) was used. After PCR, the dissociation curve was analyzed. The dissolution curve was constructed according to the manufacturer’s information provided with the ABI 7500 instrument, the relevant data were derived, and the difference between the three CT values obtained was less than 1. Data analysis was performed using Microsoft Excel 2016, and the data were calculated using the -ΔΔCt method and plotted with GraphPad Prism 5 software. Α-Tubulin was selected as a reference gene, in which the accession was MF278617.1 in the NCBI.
The real-time fluorescent quantitative PCR conditions were as follows: 94 °C for 30 s, 94 °C for 5 s, 60 °C for 15 s, and 72 °C for 10 s. Steps 2–5 required 40 to 45 cycles.
Results
Identification of the HD-Zip Gene Family Members in Larix olgensis and Analysis of the Physicochemical Properties of the Encoded Proteins
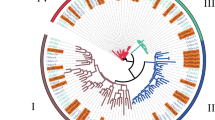
The thirteen genes were divided into four subcategories: HD-ZipI, HD-ZipII, HD-ZipIII, and HD-ZipIV. Among these genes, 5 belonged to the HD-ZipI subfamily (LoHDZ2, LoHDZ3, LoHDZ4, LoHDZ7, LoHDZ11), 2 belonged to the HD-ZipII subfamily (LoHDZ1, LoHDZ10), 4 belonged to the HD-ZipIII subfamily (LoHDZ6, LoHDZ8, LoHDZ12, LoHDZ13), and 2 belonged to the HD-ZipIV subfamily (LoHDZ5, LoHDZ9). Analysis of each subfamily revealed that the transcription factors of both LoHD-ZipI and LoHD-ZipII contained an HD region and a Zip region. The LoHD-ZipII transcription factor also contained a unique CPSCE motif consisting of 5 amino acids; in addition to the HD region and the Zip region, the LoHD-ZipIII transcription factor included a SAD region and a START region with unknown functions and a MEKHLA C-terminal domain. The LoHD-ZipIV transcription factors included an HD region, Zip region, SAD region, and START region. Furthermore, it can be seen (Fig. 2) that the conservation of the LoHD-ZipIV and LoHD-ZipIII subfamilies is higher than that of the LoHD-ZipI and LoHD-ZipII subfamilies.
HD-Zip conserved domain analysis of LoHD-ZipI~IV protein from Larix olgensis. a HD-Zip conserved domain analysis of LoHD-ZipI protein from Larix olgensis. b HD-Zip conserved domain analysis of LoHD-ZipII protein from Larix olgensis. c HD-Zip conserved domain analysis of LoHD-ZipIII protein from Larix olgensis. d HD-Zip conserved domain analysis of LoHD-ZipIV protein from Larix olgensis
The physicochemical properties of the HD-Zip transcription factor family genes in Larix olgensis showed that the length of the 13 genes is between 800 and 2500 bp and that the molecular weight of the 13 genes of the transcription factor family ranges between 31.29 and 92.72 kDa. The isoelectric point of the protein is between 4 and 7, except for that of LoHDZ1, which is within the acidic range, and the fat coefficient is higher than 50.59 (Table 4). These results indicate that the proteins of the HD-Zip transcription factor family have a relatively high thermal stability. All the average hydrophobicity coefficients are negative, which indicates that all the HD-Zip proteins are hydrophilic. According to the signal peptide analysis, the LoHD-Zip protein has no signal peptide, indicating that it is a non-secreted protein. Protein transmembrane analysis showed that all the HD-Zip proteins are non-transmembrane proteins. The subcellular localization of the 13 HD-Zip genes was predicted by SUBA4 online software. The preliminary analysis focused on the nucleus, and the results indicated that LoHD-Zip plays a role in the nucleus.
The phylogenetic tree of the HD-Zip family genes of Larix olgensis and Arabidopsis thaliana (Fig. 3) shows that HD-Zip genes have a high degree of homology between Larix olgensis and Arabidopsis. The genetic distance between LoHDZ9 and Arabidopsis AT2G032370.1 is the closest, indicating that the kinship is the closest; in addition, the genetic distance between LoHDZ11 and Arabidopsis AT3G01470.1 is close, indicating a close relationship. The results of the phylogenetic tree provides a theoretical reference for future exploration of the function of HD-Zip family genes in Larix olgensis.
The secondary structural characteristics of the 13 HD-Zip transcription factor family genes in Larix olgensis showed that the LoHD-Zip protein has α-helix (Hh), extended fold (Ee), and random coil (Cc) structures (Fig. 4, Table 5). α-Helix and random coil are the major components of the members of this family. The results showed that the secondary structures of most LoHD-ZipI, LoHD-ZipII, and LoHD-ZipIV subfamily members were mainly irregular curls and that all LoHD-ZipIII secondary structures were mainly α-helix. LoHDZ11 had the largest proportion of α-helix, while LoHDZ10 had the smallest proportion. LoHDZ10 had the largest proportion of random curl (Cc), while LoHDZ11 had the smallest proportion. LoHDZ9 had the largest proportion of extended fold (Ee), while LoHDZ3 had the smallest proportion. The protein secondary structure largely determines the protein formation configuration and is important for the function of the protein (Table 6).
Gene Expression Analysis
Expression of Genes in Different Tissues and Organs at Different Stages
Roots grown for 3 months were used as controls. The quantitative results showed that the 13 genes were expressed differently at different stages in the larch roots, stems, and leaves (Fig. 5).
LoHDZ2, LoHDZ4, LoHDZ11, and LoHDZ13 showed different degrees of downregulation in different tissues and different developmental stages compared with the control group. These results indicated that the four genes had the highest expression in non-lignified roots. The expression level of LoHDZ11 in partially lignified stems was approximately 7 times lower than that in the control group.
The expression of LoHDZ5, LoHDZ9, and LoHDZ10 showed different degrees of upregulation in the treatment groups compared with the control group. LoHDZ5 was highly expressed in partially lignified and fully lignified leaves. The expression level of LoHDZ9 in fully lignified roots and non-lignified leaves was approximately 6 times higher than that in the control group, and LoHDZ10 had a relatively high expression level in the leaves.
LoHDZ8 had the highest expression level in the partially lignified roots, and the expression level was approximately 10 times higher than that in other organs.
Gene Expression Under Drought Stress
All LoHD-Zip genes were significantly differentially regulated under different concentrations of PEG6000 (Fig. 6).
The expression of LoHDZ1 and LoHDZ5 was upregulated in response to various concentrations of PEG6000 for different times. The expression level was the highest after treatment with 28% PEG6000 for 48 h, which was approximately 10 times higher than that of the control group.
LoHDZ3, LoHDZ4, and LoHDZ13 were downregulated in response to various concentrations of PEG6000 treatment. The expression level of LoHDZ3 was downregulated after treatment with 21% PEG6000 for 24 h and 28% PEG6000 for both 72 h and 96 h, the latter of which was approximately 10 times higher than that of the control group. The expression level of LoHDZ4 was the lowest after treatment with 7% and 14% PEG6000 for 96 h. After treatment with 21% PEG6000, the expression level of LoHDZ4 was the lowest but was not significantly different after 72 h or 96 h, and the expression level was the lowest after treatment with 28% PEG6000 for 72 h. LoHDZ13 had the lowest expression level after treatment with 14% PEG6000 for 48 h and 21% PEG6000 for 24 h.
The expression levels of LoHDZ2 and LoHDZ7 after 21% and 28% PEG6000 treatments (except the 21% PEG6000 treatment for 24 h) were lower than those in the control group. In response to treatment with 21% PEG6000 for 96 h, the expression level was especially the lowest; LoHDZ10 was downregulated in all time periods under the 14%, 21%, and 28% PEG6000 treatments. The expression level after 14% PEG6000 treatment for 72 h was approximately 13 times lower than that of the control group.
Gene Expression Under Salt Stress
The expression levels of the 13 LoHD-Zip genes in larch seedlings treated with 0.2 M NaCl solution showed that each gene was downregulated after salt stress treatment, and the expression of the 13 genes was the lowest after 96 h of treatment (Fig. 7). The expression level of 5 genes (LoHDZ1, LoHDZ2, LoHDZ7, LoHDZ8, LoHDZ11) was approximately 5 times lower than that of the control group after 96 h of treatment. In addition, the expression levels of 6 genes (LoHDZ3, LoHDZ4, LoHDZ5, LoHDZ6, LoHDZ9, LoHDZ13) after treatment for 96 h were approximately 8 times lower than those of the control group.
Discussion
Identification and Analysis of the Physicochemical Properties of the HD-Zip Family Genes in Larch
In recent years, genome-wide identification of HD-Zip family genes in various plant species has been performed. For example, researchers have identified 50, 33, and 63 HD-Zip genes in the model plant species Arabidopsis (Ciarbelli et al. 2008), rice (Agalou et al. 2008), and poplar (Hu et al. 2012), respectively. In addition, the functions of HD-Zip family genes at the whole-genome level have been studied in plant species such as soybean (Belamkar et al. 2014), wheat (Yue et al. 2018), corn (Zhao et al. 2011), and cassava (Ding et al. 2017), including their expression patterns under abiotic stresses such as high salt, drought, chilling injury, and ABA. These functions serve as a certain reference basis for HD-Zip family genes in terms of their regulation of plant abiotic stress responses. However, larch HD-Zip family genes have not yet been investigated in this respect.
Based on the results of the transcriptome sequencing of Larix olgensis, 32 candidate sequences with HD domains were identified (Table 7). When the Arabidopsis HD-Zip family sequences were used as a basis for comparison, 13 HD-Zip genes with an intact CDS domain were identified; this number was smaller than that of other investigated species. The isoelectric points of the four subfamilies of the larch HD-Zip family genes are mostly acidic, mainly because the proteins contain many acidic amino acids, including serine. The fat coefficient is greater than 50.59, and most of the proteins are highly hydrophilic. The secondary structure prediction results show that α-helix and random coil account for the largest proportion, and members of the same subfamily exhibit similar trends, as in a study in birch (Prigge and Clark 2006). Therefore, these findings provided further evidence for identifying HD-Zip family genes in Larix olgensis. Although the secondary structure of each subfamily of genes is similar, their tertiary structure is different. The differences and similarities in the spatial structure of these proteins may lead to functional similarities and differences. The results of the signal peptide analysis showed that HD-Zip proteins did not have a signal peptide, indicating that they are non-secreted proteins, and the results of the protein transmembrane analysis revealed that the HD-Zip proteins were non-transmembrane proteins. It is speculated that the properties of these proteins may be related to the functions of these protein families. The results of this study therefore provide a reference for future studies of the functions of this family in larch.
Tissue-Specific Expression of LoHD-Zip Genes
HD-Zip transcription factors are present in low-lying bryophytes from ferns and gymnosperms to more evolved angiosperms. These transcription factor family genes play an important regulatory role in the growth and development of plants (Floyd 2006). In Arabidopsis, rice, and Populus euphratica, HD-ZipIII genes are strongly expressed in the vascular bundles of roots, stems, and leaves. Localization revealed that HD-ZipIII genes in Arabidopsis thaliana are specifically expressed in protoplasts and xylem cells (Kangj and Denglar 2002). This study preliminarily targeted the expression patterns of members of the HD-Zip family in the roots, stems, and leaves of Larix olgensis during the periods of complete lignification, partial lignification, and unlignification. Real-time fluorescent quantitative PCR was performed on these 13 genes. The results showed that 8 of them (LoHDZ2, LoHDZ4, LoHDZ5, LoHDZ8, LoHDZ9, LoHDZ10, LoHDZ11, LoHDZ13) were expressed specifically in different tissues at different stages.
According to previous studies, members of the HD-ZipI subfamily are mainly involved in light signal transduction, abiotic stress, and leaf development processes (Wei et al. 2018). Members of the HD-ZipI subfamily ATHB1 in Arabidopsis are a transcriptional activator involved in leaf development (Aoyama et al. 1995). LoHDZ2, LoHDZ4, and LoHDZ11 belong to the HD-ZipI subfamily. The results showed that these three genes had the highest expression in non-lignified roots. However, the LoHD-ZipI subfamily gene expression level in the non-lignified roots was higher than that in the leaves, suggesting that the members of this subfamily may be involved in root development, which is the same conclusion as that from Xiuming’s research on tomato HD-Zip family genes (Xiuming 2017).
In Arabidopsis, HD-ZipII subfamily genes were mainly involved in environmental responses, the light quality response, the shade avoidance response, and abiotic stress responses (Wei et al. 2018; Xiuming 2017). Light of different qualities or wavelengths has distinct biological effects, including different effects on plant morphological structure and chemical composition, photosynthesis, and organ growth and development. According to a previous study, the Arabidopsis HD-ZipII subfamily gene ATHB17 played a role in regulating leaf morphology, chlorophyll content, chloroplast number, and photosynthesis (Hymus et al. 2013). LoHDZ10 belongs to the HD-ZipII subfamily. According to the evolutionary phylogenetic tree in this study, LoHDZ10 clustered with two genes of the Arabidopsis HD-ZipII subfamily. The expression level of LoHDZ10 in the leaves was significantly higher than that in the stems and roots, indicating that it may be involved in leaf development, which is consistent with results of Arabidopsis research.
In Arabidopsis studies, the HD-ZipIII protein was shown to control the establishment of the various polarities (including the polarity of leaves and stem axes), paraxial cells, paraxial structure, and leaf and vascular system formation; regulate the meristematic growth of stem tips; and promote the formation of meristems and both the proliferation and differentiation of stem cells (Henriksson 2005). Baima et al. (2001) found that the overexpression of the HD-ZipIII subfamily gene ATHB-8 in Arabidopsis increased xylem tissue production and overexpression in leaf and root protoplast cells. LoHDZ8 and LoHDZ13 belong to the HD-ZipIII subfamily. The expression of LoHDZ8 in partially lignified roots was relatively high, but the expression of LoHDZ13 in non-lignified roots was higher, indicating that these two genes may be involved in root development and that their function may be similar to that of Arabidopsis HD-ZipIII subfamily genes.
The HD-ZipIV subfamily genes mainly participate in the differentiation and material accumulation within epidermal and subepithelial cells (Baima et al. 2001). According to previous studies, the Arabidopsis HD-ZipIV subfamily gene ANL2 was involved in the regulation of leaf epidermal anthocyanin accumulation and root cell differentiation (Harris et al. 2011). The expression level of LoHDZ5 in partially lignified and fully lignified leaves was relatively high; moreover, LoHDZ9 was highly expressed in fully lignified roots and non-lignified leaves. LoHDZ5 and LoHDZ9 belong to the HD-ZipIV subfamily, and it was speculated that LoHD-ZipIV subfamily genes also may be involved in the development of roots and leaves of Larix olgensis.
Analysis of Differentially Expressed LoHD-Zip Genes Under Abiotic Stress
Studies had shown that the expression of HD-Zip transcription factor I and II subfamily genes was induced by drought, high salt, ABA, and chilling. These two subfamily genes are involved in the hormone signaling pathway and regulate plant cell expansion, division, and differentiation to enhance plant tolerance by interacting with hormone pathway genes and other downstream genes (Harris et al. 2011). Current studies on HD-Zip transcription factor I and II subfamilies had focused on Arabidopsis, rice, and sunflower (Agalou et al. 2008). Studies on the Arabidopsis HD-ZipI subfamily suggest that ATHB7 played a role in signaling pathways that mediate the drought responses (Soderman et al. 1996); ATHB12 and ATHB7 had similar molecular mechanisms, as these genes played a role in the regulation of water-deficient growth responses (Olsson et al. 2004). In this study, two-month-old Larix olgensis seedlings were treated with solutions of PEG6000 at concentrations of 7%, 14%, 21%, and 28%. The results showed that LoHDZ2, LoHDZ3, LoHDZ4, and LoHDZ7 of the HD-ZipI subfamily showed obvious downregulated expression. Genes within the same subfamily had the same conserved domain and similar protein secondary and tertiary structures, so they may have similar functions. Accordingly, it is speculated that the HD-ZipI subfamily of Larix olgensis may also be involved in the response to drought stress expression patterns; the expression of the HD-ZipII subfamily gene LoHDZ1 was upregulated under drought stress. Similarly, the Arabidopsis HD-ZipII subfamily gene ATHB17 also showed hypersensitivity to drought stress in young seedlings (Park et al. 2013). Moreover, it can be seen from the phylogenetic tree that LoHDZ1 and LoHDZ2 clustered together, indicating that their homology is high, and it is speculated that they may have similar functions. Therefore, LoHDZ1 may also be involved in drought stress responses. Additionally, the expression of the LoHDZ5 of the HD-ZipIV subfamily was upregulated at various time points in the different treatments. Yu et al. (2008) showed that the three genes of the Arabidopsis HD-ZipIV subfamily could improve drought tolerance, and Fu et al. (2013) found that the overexpression of HDG11 was positively correlated with the drought tolerance of plants and the activity level of SOD. It is speculated that LoHDZ5 also played an important role in the response to drought stress.
According to previous studies, Nakamura et al. (2006) significantly induced the expression of the Arabidopsis HD-ZipI genes AtHB7 and AtHBl2 with 100 mmol/L NaCl, and the expression levels were upregulated by 12 and 25 times; however, the expression levels of the MH381401 and MH381603 genes in Betula platyphylla were slightly downregulated after salt stress treatment (Wang et al. 2018). AtHDG1 from Arabidopsis can also improve the salt tolerance of cotton, poplar, and perennial grass species (Cao et al. 2009). Oshox22 in rice affects ABA biosynthesis and regulates drought and salt responses, which indicates involvement of this gene in ABA-mediated signal transduction pathways (Zhang et al. 2012); similarly, the Zmhdz10 gene in maize can actively regulate the drought tolerance and salt tolerance of plants via ABA-dependent signaling pathways (Zhao et al. 2014a, b), and Hahb4 in sunflower responded to high salt, drought, and abiotic stresses such as ABA (Dezar et al. 2005). In this study, the expression of 13 genes showed significant downregulation at 96 h after treatment with 0.2 M NaCl solution. Consistent with the conclusions in the above studies, it is speculated that the expression of the 13 genes of the family in the present study is induced by salt, and the specific regulatory mechanism remains to be studied. Based on the existing transcriptome data of Larix olgensis, this study identified and analyzed the larch HD-Zip family and provided a basis for future studies of these gene family members and their function in Larix olgensis.
Availability of Data and Material
Reliable and transparent data.
References
Agalou A, Purwantomo S, Overnas E et al (2008) A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol Biol 66(1–2):87–103
Aoyama T, Dong CH, Wu Y et al (1995) Ectopic expression of the Arabidopsis transcriptional activator Athb-1 alters leaf cell fate in tobacco. Plant Cell 7(11):1773–1785
Ariel FD, Manavella PA, Dezar CA, Chan RL (2007) The true story of the HD-Zip family. Trends Plant Sci 12(9):419–426
Baima S, Possenti M, Matteucci A, Wisman E, Altamura MM, Ruberti I, Morelli G (2001) The Arabidopsis ATHB-8 HD-zip protein acts as a differentiation-promoting transcription factor of the vascular meristems. Plant Physiol 126(2):643–655
Belamkar V, Weeks NT, Bharti AK, Farmer AD, Graham MA, Cannon SB (2014) Comprehensive characterization and RNA-Seq profiling of the HD-Zip transcription factor family in soybean (Glycine max) during dehydration and salt stress. BMC Genomics 15:950
Bharathan G, Janssen BJ, Kellogg EA, Sinha N (1997) Did homeodomain proteins duplicate before the origin of angiosperms fungi and metazoa? Proc Natl Acad Sci U S A 94(25):13749–13753
Cao YJ, Wei Q, Liao Y, Song HL, Li X, Xiang CB, Kuai BK (2009) Ectopic overexpression of AtHDG11in tall fescue resulted in enhanced tolerance to drought and salt stress. Plant Cell Rep 28(4):579–588
Carabelli M, Turchi L, Ruzza V et al (2013) Homeodomain-Leucine zipper II family of transcription factors to the limelight:central regulators of plant development
Ciarbelli AR, Ciolfi A, Salvucci S, Ruzza V, Possenti M, Carabelli M, Fruscalzo A, Sessa G, Morelli G, Ruberti I (2008) The Arabidopsis Homeodomain-leucine Zipper II gene family:diversity and redundancy. Plant Mol Biol 68(4–5):465–478
Dezar CA, Fedrigo GV, Chan RL (2005) The promoter of the sunflower HD-zip protein gene Hahb4 directs tissue-specific expression and is inducible by water stress, high salt concentrations and ABA. Plant Sci 169(2):0–456
Ding ZH, Fu LL, Yan Y et al (2017) Genome-wide characterization and expression profiling of HD-Zip gene family related to abiotic stress in cassava. PLoS One 12(3):e0173043
Floyd SK (2006) Evolution of class III homeodomain-leucine zipper genes in streptophytes. Genetics 173(1):373–388
Franklin KA, Praekelt U, Stoddart WM, Billingham OE, Halliday KJ, Whitelam GC (2003) Phytohromes B,D and E act redundantly to control multiple physiological responses in Arabidopsis. Plant Physiol 131(3):1340–1346
Fu R, Liu W, Li Q et al (2013) Comprehensive analysis of the homeodomain-leucine zipper IV transcript. Genome 56(7):395–405
Harris JC, Hrmova M, Lopato S, Langridge P (2011) Modulation of plant growth by HD-Zip class I and II transcription factors in response to environmental stimuli. New Phytol 190(4):823–837
Harris JC, Sornaraj P, Taylor M, Bazanova N, Baumann U, Lovell B, Langridge P, Lopato S, Hrmova M (2016) Molecular interactions of the y-clade homeodomain -leucine zipper class I transcription factors during the wheat response to water deficit. Plant Mol Biol 90(4–5):435–452
Henriksson E (2005) Homeodomain leucine zipper class I genes in Arabidopsis. Expression patterns and phylogenetic relationships. Plant Physiol 139(1):509–518
Henriksson E, Olsson ASB, Johannesson H, Johansson H, Hanson J, Engström P, Söderman E (2005) Homeodomain leucine zipper class I genes in Arabidopsis expression patterns and phylogenetic relationships. Plant Physiol 139(1):509–518
Hu RB, Chi XY, Chai GH et al (2012) Genome-wide identification,evolutionary expansion and expression profile of homeodomain-leucine zipper gene family in poplar (Populus trichocarpa). PLoS One 7(2):e31149
Hymus GJ, Cai S et al (2013) Application of HB17,an Arabidopsis class II homeodomain-leucine zipper transcription factor to regulate chloroplast number and photosynthetic capacity. J Exp Bot 64(14):4479–4490
Ingram GC, Boisnard-Lorig C, Dumas C, Rogowsky PM (2000) Expression patterns of genes encoding HD-Zip homeo domain proteins define specific domains in maize embryos and meristems. Plant J 22(5):401–414
Ito M, Sentoku N, Nishimura A, Hong SK, Sato Y, Matsuoka M (2002) Position dependent expression of GL2-type homeobox gene Rocl:significance for protoderm differentiation and radial pattern formation in early rice embryogenesis. Plant J 29(4):497–507
Jain M, Tyagi AK, Khurana JP (2008) Genome-wide identification, classification, evolutionary expansion and expression analyses of homeobox genes in rice. FEBS J 275(11):2845–2861
Jinhua L et al (2003) Current status and development strategies of wood fiber raw materials for pulp and papermaking in China. CSIR 16(6):32–35
Kangj, Denglar N (2002) Cell cycling frequency and expression of the homeobox gene ATHB-8 during leaf vein development in Arabidopsis. Planta 216(2):212–219
Kim YK, Son O, Kim MR, Nam KH, Kim GT, Lee MS, Choi SY, Cheon CI (2007) ATHB2, an Arabidopsis class I homeodomain-leucine zipper gene, is expressed in the adaxial region of young leaves. Plant Cell Rep 26(8):1179–1185
Lian L, Xu H et al (2019) Expression of antioxidant enzyme genes in rice under PEG-simulated drought-stress. FJAS 34(3):255–263
Liu L, Hai DU, Tang XF et al (2008) The roles of MYB transcription factors on plant defense responses and its molecular mechanism. Hereditas 30(10):1265–1271
Ma Q, Kang J, Long R, Zhang T, Xiong J, Zhang K, et al. (2017). Comparative proteomic analysis of alfalfa revealed new salt and drought stress-related factors involved in seed germination. Molecular Biology Reports 44:261–170
Mukherjee K, Brocchieri L, Burglin TR (2009) A comprehensive classification and evolutionary analysis of plant homeobox genes. Mol Biol Evol 26(12):2775–2794
Nakamura M, Katsumata H, Abe M, Yabe N, Komeda Y, Yamamoto KT, Takahashi T (2006) Characterization of the class IV homeodomain-leucine zipper gene family in Arabidopsis. Plant Physiol 141(4):1363–1375
Olsson A, Engström P, Söderman E (2004) The homeobox genes ATHB12 and ATHB7 encode potential regulators of growth in response to water deficit in Arabidopsis. Plant Mol Biol 55(5):663–677
Park MY, Kim SA, Lee SJ, Kim SY (2013) ATHB17 is a positive regulator of abscisic acid response during early seedling growth. Mol Cell 35(2):125–133
Prigge MJ, Clark SE (2006) Evolution of the class III HD-Zip gene family in land plants. Evol Dev 8(4):350–361
Prigge MJ, Otsuga D, Alonso JM, Ecker JR, Drews GN, Clark SE (2005) Class III homeodomain-leucine zipper gene family members have overlapping, antagonistic,and distinct roles in Arabidopsis development. Plant Cell 17(1):61–76
Robischon M, Du J, Miura E et al (2011) The Populus classIII HD-ZIP popREVOLUTA influences cambium initiation and patterning of woody stems. Plant Physiol 55(3):1214–1225
Schena M, Davis RW (1992) HD-Zip proteins:members of an Arabidopsis homeodomain protein superfamily. Proc Natl Acad Sci U S A 89(9):3894–3898
Soderman E, Mattsson J, Engstrm P (1996) The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J 10(2):375–381
Wang J, Zhang W, Li L (2018) Bioinformatics and response salt stress analysis of HD-Zip gene family in Betula platyphylla. Plant Res 38(06):931–938
Wei L, Yi W, Wang K, Zhao M et al (2018) The structure, function and expression pattern of HD-Zip gene family. Bulletin of Botanical Research 16(03):781–790
Xin-Yu Z, Yan-Ning G (2004) To design PCR primers with Oligo 6 and Primer Premier 5. Bioinformatiocs
Xiuming J (2017) Bioinformatics analysis of tomato HD-Zip transcription factor family and identification of HD-ZipI subfamily stress-resistance related genes. Northeast Agricultural University
Yu X, Bao F (2010) Study on the sharpness equation of larch plantation. Northeast Forestry University
Yu H, Chen X, Yuan-Yuan et al (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20(4):1134–1151
Yue H, Shu DT, Wang M et al (2018) Genome-wide identification and expression analysis of the HD-Zip gene family in wheat (Triticum aestivum L). Genes 9(2):70
Zhang S, Haider I, Kohlen W, Jiang L, Bouwmeester H, Meijer AH, Schluepmann H, Liu CM, Ouwerkerk PBF (2012) Function of the HD-Zip I gene Oshox22 in ABA-mediated drought and salt tolerances in rice. Plant Mol Biol 80(6):571–585
Zhang X, Yao W, Zhao K, Jiang T, Zhou B (2017) Analysis of bioinformatics and response salt stress in HD-Zip transcription factor family of Populus nigra. Plant Res 37(05):715–721
Zhao Y, Zhou YQ, Jiang HY et al (2011) Systematic analysis of sequences and expression patterns of drought-responsive members of the HD-Zip gene family in maize. PLoS One 6(12):e28488
Zhao Y, Ma Q, Xiaolei et al (2014a) A novel maize homeodomain-leucine zipper (HD-Zip) I gene, Zmhdz10, positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol 55(6):1142–1156
Zhao Y, Ma Q, Jin XL et al (2014b) A novel maize homeodomain-leucine zipper HD-Zip I gene Zmhdzl0 ,positively regulates drought and salt tolerance in both rice and Arabidopsis. Plant Cell Physiol 55(6):1142–1156
Zhu YY, Song DL, Sun JY et al (2013) PtrHB7,a classIII HD-Zip gene, plays a critical role in regulation of vascular cambium differentiation in Populus. Mol P1ant 6(4):1331–1134
Funding
This project was supported by the National Science and Technology Major Project (2018ZX08020003-001-001), the National Natural Science Foundation of China (Grant No.31700595), and the Fundamental Research Funds for the Central Universities (2572019BA13), Heilongjiang Touyan Innovation Team Program.
Author information
Authors and Affiliations
Contributions
Peiqi An and Lei Zhang conceived and designed the study. Peiqi An, Qing Cao, and Chen Wang performed the experiments. Peiqi An wrote the paper. Junhun Wang, Hanguo Zhang, and Lei Zhang reviewed and edited the manuscript. All authors read and approved the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethics Aapproval
Not applicable.
Code Availability
Not applicable.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key Message
In this study, we screened and identified thirteen HD-Zip family genes with complete CDS regions based on the sequencing results of Larix olgensis. According to the characteristics, we divided these genes into four subfamilies: HD-ZipI, HD-ZipII, HD-ZipIII, and HD-ZipIV. We used bioinformatics-related software and online websites to analyze the physiological and biochemical indexes and protein structure of HD-Zip family genes. And we performed real-time fluorescence quantitative analysis of these genes in different tissues at different periods and in response to abiotic stress.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
An, P., Cao, Q., Wang, C. et al. Spatiotemporal Expression and Bioinformatic Analyses of the HD-Zip Transcription Factor Family in Larix olgensis. Plant Mol Biol Rep 39, 212–225 (2021). https://doi.org/10.1007/s11105-020-01244-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-020-01244-9