Abstract
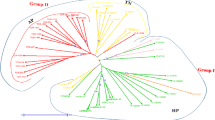
Development of genomic resources in any crop is the pre-requisite for the construction of linkage map and implementation of molecular breeding strategies to develop superior cultivars. Large number of molecular markers are required to enrich the scanty information available in horsegram (Macrotyloma uniflorum).We employed the next-generation Illumina sequencing platform to develop a large number of microsatellite markers in this species. Of the total 23,305 potential SSRs motifs, 5755 primers were designed. Of these, 1425, 1310, 856, 1276, and 888 were of di-, tri-, tetra-, penta-, and hexa-nucleotide repeats respectively. Thirty polymorphic SSR primers and 24 morphological traits were used in 360 horsegram accessions to detect the genetic diversity and population structure. Thirty primers amplified 170 polymorphic alleles with an average of 5.6 alleles per primer having size 80 to 380 bp. The polymorphism information content (PIC) ranged from 0.15 to 0.76 with an average of 0.50, suggesting that SSR markers used in the study were polymorphic and suitable for characterization of horsegram germplasm. Dendrogram-based on Jaccard’s similarity coefficient and neighbor-joining tree grouped the horsegram accessions into two major clusters. Similarly, STRUCTURE analysis assigned genotypes into two gene pools namely Himalayan origin and Southern India. Diversity analysis based on 24 agro-morphological traits also suggested the presence of high level of diversity among the accessions.





Similar content being viewed by others
References
Arora RK, Chandel KPS (1972) Botanical source area of wild herbage legumes in India. Trop grassland 6:213–221
Bhartiya A, Aditya JP, Kant L (2015) Nutritional and remedial potential of an underutilized food legume horsegram (Macrotyloma uniflorum): a review. J Ani Plant Sci 25:908–920
Bhardwaj J, Chauhan R, Swarnkar MK, Chahota RK, Singh AK, Shankar R, Yadav SK (2013) Comprehensive transcriptomic study on horsegram (Macrotyloma uniflorum): de novo assembly, functional characterization and comparative analysis in relation to drought stress. BMC Genomics 14:647–665
Bhatt R, Karim AA (2009) Exploring the nutritional potential of wild and underutilized legumes. Comp Rev Food Sci Food Saf 8:305–331
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphism. Am J Hum Genet 32:314–331
Chahota RK, Sharma TR, Dhiman KC, Kishore N (2005) Characterization and evaluation of Horsegram (Macrotyloma uniflorum Roxb.) germplasm for Himachal Pradesh. Indian J Pl ant Genet Resour 18:221–223
Chahota RK, Sharma TR, Sharma SK, Kumar N, Ran JC (2013) Horsegram: in: Mohar Singh, Hari D. Upadhyaya and Ishwari Singh Bisht (eds) genetic and genomic resources of grain legume improvement, Elsevier insight pp 293–305
Chaitanya DAK, Santosh Kumar M, Reddy AM, Mukherjee NSV, Sumanth MH, Ramesh DA (2010) Antiurolithiatic activity of Macrotyloma uniflorum seed extraction ethylene glycol induced urolithiasis in albino rats. J Innov Tre Pharm Sci 1:216–226
Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127-1309–1321
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure from small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Durga KK (2012) Variability and divergence in horsegram (Dolichos uniflorus). J Arid Land 4:71–76
Earl DA, Bridgett M, VonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:259–261
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Gopalan C, Ramashastri BV, Balasubramanyan SC (1999) Nutritive value of Indian foods. Hyderabad, India: National Institute of Nutrition, ICMR pp 156
Jayaraj AP, Tovey FI, Lewin MR, Clark GC (2000) Duodenal ulcer prevalence: experimental evidence for possible role of dietary lipids. J Gastroenterol Hepatol 15:610–616
Kofler R, Schlotterer C and Lelley T (2007) SciRoKo: a new tool for whole genome microsatellite search and investigation. Bioinformatics.1:1683–1685
Kulkarni GB (2010) Evaluation of genetic diversity of horsegram (Macrotyloma uniflorum) germplasm through phenotypic trait analysis. Green Farm 1:563–565
Neelam S, Kumar V, Natarajan S, Venkateshwaran K, Pandravada SR (2014) Evaluation and diversity observed in Horsegram (Macrotyloma uniflorum (lam) Verdc.) germplasm from Andhra Pradesh, India. Int J Plant Res 4:17–22
Latha M, Suma A, Asha KI, Dwivedi NK, Mani S, Indiradevi A (2013) Seed polymorphism in south Indian horsegram (Macrotyloma uniflorum (lam.) Verdc.): a comprehensive study. J appl biol biotechnol 1:001–006
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, Li S, Yang H, Wang J, Wang J (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Luikart G, England PR, Tallmon D, Jordan S, Taberlet P (2003) The power and promise of population genomics: from genotyping to genome typing. Nat Rev Genet 4:981–994
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with non- repetitive DNA in plant genomes. Nat Genet 30:194–200
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Perrier X, Jacquemoud-Collet JP (2006) DARwin software. http://Darwin. Cirad. Fr/Darwin
Rozen S, Skaletsky HJ (2000) Primer3 for general users and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, pp 365–386
Ramesh CK, Rehman A, Prabhakar BT, Vijay ABR, Aditya SJ (2011) Antioxidant potential in sprout vs. seeds of Vigina radiate and Macrotyloma uniflorum. J App Pharm Sci 1:99–103
Rick CM, Holle M (1990) Andean Lycopersicum esculentum var. cerasiforme: genetic Varaition and its evolutionary significance. Econ Bot 44:69–78
Sharma V, Sharma TR, Rana JC, Chahota RK (2015a) Analysis of genetic diversity and population structure in Horsegram (Macrotyloma uniflorum) using RAPD and ISSR. Agric Res 4(3):221–230
Sharma V, Rana M, Katoch M, Sharma PK, Ghani M, Rana JC, Sharma TR, Chahota RK (2015b) Development of SSR and ILP markers in horsegram (Macrotyloma uniflorum), their characterization, cross-transferability and relevance for mapping. Mol Breed 35:102
Sodani SN, Paliwal RV, Jain LK (2004) Phenotypic stability for seed yield in rain fed horse gram (Macrotyloma uniflorum). Proceedings of the National Symposium on arid legumes for sustainable agriculture and trade; jodhpur, Rajasthan, India: central arid zone research institute pp
Verdcourt B (1971) Phaseoleae. In: Gillet JB, Polhill RM, Verdcourt B (eds) Flora of east tropical Africa. Leguminosae subfamily Papilionoideae. Crown Agents 2:581–594
Varma VS, Durga KK, Ankaiah R (2013) Assessment of genetic diversity in horsegram (Dolichos uniflorus). EbSCO online library
Venkatesha RT (1999) Studies on molecular aspects of seed storage proteins in horsegram (Dolichos fiblorus L.) Ph.D. thesis. Department of Food microbiology Central Food Technological Research Institute, Mysore, India
Yadav S, Negi KS, Mandal S (2004) Protein and oil rich wild horsegram. Genet Resour Crop Ev 51:629–633
Williams CE, St. Clair DA (1993) Phenetic relationships and levels of variability detected by restriction fragment length polymorphism and random ampliWed polymorphic DNA analysis of cultivated and wild accessions of Lycopersicon esculentum. Genome 36:619–630
Yeh FC, Boyle TJB (1997) Population genetic analysis of codominant and dominant markers and quantitative traits. Belg Journ Bot 129:157
Zhang S, Tang C, Zhao Q, Li J, Yang L, Qie L, Fan X, Li L, Zhang N, Zhao M, Liu X, Chai Y, Zhang X, Wang H, Li Y, Li W, Zhi H, Jia G, Diao X (2014) Development of highly polymorphic simple sequence repeat markers using genome-wide microsatellite variant analysis in foxtail millet [Setaria italica (L.) P. Beauv.] BMC genomics: DOI: 10.1186/1471-2164-15-78
Acknowledgements
This work was funded by the Department of Science and Technology, Govt of India, and Japan Society for the Promotion of Science, Japan in the form of Collaborative Research Project.
Author Contribution Statement
RKC and TRS designed the experiments and wrote the manuscript. JCR provided the material. DS and MR performed the experiment. SI and HK designed the primers from Illumina sequence data. VS and AN analyzed data.
Key Message
This is the first attempt to develop the genomic SSR markers in this crop and it is also a maiden attempt to assess the genetic diversity using molecular and morphological markers in lesser known but important legume. The study revealed the presence of two gene pools Indian horsegram germplasm.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Ethical Standards
The authors declare that the experiments comply with the current laws of the countries in which the experiments were performed
Electronic supplementary material
Supplementary Fig 1
Distribution of horsegram germplasm in India (DOC 355 kb)
Supplementary Fig 2
Population structure of 360 horsegram accessions showing two genetic stocks (DOC 39 kb)
Table 1
(DOCX 136 kb)
Table 2
(DOCX 272 kb)
Table 3
(DOCX 233 kb)
Table 4
(DOCX 279 kb)
Table 5
(DOCX 227 kb)
Table 6
(DOCX 18 kb)
Table 7
(DOCX 16 kb)
Rights and permissions
About this article
Cite this article
Chahota, R.K., Shikha, D., Rana, M. et al. Development and Characterization of SSR Markers to Study Genetic Diversity and Population Structure of Horsegram Germplasm (Macrotyloma uniflorum). Plant Mol Biol Rep 35, 550–561 (2017). https://doi.org/10.1007/s11105-017-1045-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-017-1045-z