Abstract
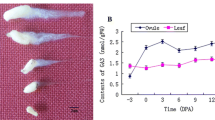
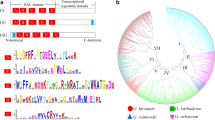
DELLA proteins act as repressors in the gibberellin (GA) signaling pathway. GA could relieve this repression by triggering DELLA protein degradation via the ubiquitin–proteasome pathway. In this study, we cloned four genes from allotetraploid cotton that encode DELLA-like proteins whose functions have not been characterized previously. These four genes were designated as GhGAI3a, GhGAI3b, GhGAI4a, and GhGAI4b. The deduced proteins encoded by these genes contain all domains present in typical DELLA proteins. Sequence analysis showed that GhGAI3a belongs to the D sub-genome, whereas GhGAI3b belongs to the A sub-genome. The results from the analysis of their expression patterns in allotetraploid cotton showed that their expression was regulated during the fiber initiation and elongation stages. GhGAI3a was expressed at a high level at the fiber initiation stage; however, its transcript level was significantly reduced in fiber cells during the early fiber elongation stage. To test the function of GhGAI3a, we generated transgenic Arabidopsis lines that carried a construct in which expression of GhGAI3a was under the control of the cauliflower mosaic virus 35S promoter. The transgenic Arabidopsis lines exhibited smaller leaves and shorter stems and stamens. In addition, the GhGAI3a overexpression lines showed a defect in GA-induced hypocotyl elongation. These results suggest that GhGAI3a acts as a repressor of the GA signaling pathway.






Similar content being viewed by others
References
Alabadi D, Gil J, Blazquez M, Garcia-Martinez J (2004) Gibberellins repress photomorphogenesis in darkness. Plant Physiol 134:1050–1057
Aleman L, Kitamura J, Abdel-mageed H, Lee J, Sun Y, Nakajima M, Ueguchi-Tanaka M, Matsuoka M, Allen RD (2008) Functional analysis of cotton orthologs of GA signal transduction factors GID1 and SLR1. Plant Mol Biol 68:1–16
Barthelson RA, Qaisar U, Galbraith DW (2010) Functional analysis of the Gossypium arboreum genome. Plant Mol Biol Rep 28:334–343
Beasley CA, Ting IP (1974) The effects of plant growth substances on in vitro fiber development from unfertilized cotton ovules. Am J Bot 61:188–194
Bolle C (2004) The role of GRAS proteins in plant signal transduction and development. Planta 218:683–692
Boss PK, Thomas MR (2002) Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature 416:847–850
Chandler PM, Marion-Poll A, Ellis M, Gubler F (2002) Mutants at the Slender1 Locus of Barley cv Himalaya. Molecular and physiological characterization. Plant Physiol 129:181–190
Chen JG, Du XM, Zhao HY, Zhou X (1996) Fluctuation in levels of endogenous plant hormones in ovules of normal and mutant cotton during flowering and their relation to fiber development. J Plant Growth Regul 15:173–177
Fleet DM, Sun TP (2005) A DELLAcate balance: the role of gibberellin in plant morphogenesis. Curr Opin Plant Biol 8:77–85
Foster T, Kirk C, Jones WT, Allan AC, Espley R, Karunairetnam S, Rakonjac J (2007) Characterisation of the DELLA subfamily in apple (Malus x domestica Borkh.). Tree Genet Genomes 3:187–197
Gokani SJ, Thaker VS (2002) Role of gibberellic acid in cotton fibre development. J Agric Sci 138:255–260
Hussain A, Cao D, Cheng H, Wen Z, Peng J (2005) Identification of the conserved serine/threonine residues important for gibberellin-sensitivity of Arabidopsis RGL2 protein. Plant J 44:88–99
Ikeda A, Ueguchi-Tanake M, Sonoda Y, Kitano H, Koshioka M, Futsuhara Y, Matsuoka M, Yamaguchi J (2001) Slender rice, a constitutive gibberellin response mutant, is caused by a null mutation of the SLR1 gene, an ortholog of the height-regulating gene GAI/RGA/RHT/D8. Plant Cell 13:999–1010
Itoh H, Ueguchi-Tanaka M, Sato Y, Ashikari M, Matsuoka M (2002) The gibberellin signaling pathway is regulated by the appearance and disappearance of SLENDER RICE1 in nuclei. Plant Cell 14:57–70
Ji SJ, Lu YC, Feng JX, Wei G, Li J, Shi YH (2003) Isolation and analyses of gene preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res 31:2534–2543
John ME, Crow LJ (1992) Gene expression in cotton (Gossypium hirsutum L.) fiber: cloning of the mRNAs. Proc Natl Acad Sci 89:5769–5773
Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J (2002) Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev 16:646–658
Lee JJ, Woodward AW, Chen ZJ (2007) Gene expression changes and early events in cotton fibre development. Ann Bot 100:1391–1401
Li XB, Fan XP, Wang XL, Cai L, Yang WC (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17:859–875
Li Y, Liu D, Tu L, Zhang X, Wang L, Zhu L, Tan J, Deng F (2010) Suppression of GhAGP4 gene expression repressed the initiation and elongation of cotton fiber. Plant Cell Rep 29:193–202
Liao WB, Ruan MB, Cui BM, Xu NF, Lu JJ, Peng M (2009) Isolation and characterization of a GAI/RGA-like gene from Gossypium hirsutum. Plant Growth Regul 58:35–45
Luo M, Xiao Y, Li X, Lu X, Deng W, Li D, Hou L, Hu M, Li Y, Pei Y (2007) GhDET2, a steroid 5a-reductase, plays an important role in cotton fiber cell initiation and elongation. Plant J 51:419–430
Meng XP, Li FG, Liu CL, Zhang CJ, Wu ZX, Chen YJ (2010) Isolation and characterization of an ERF transcription factor gene from cotton (Gossypium barbadense L.). Plant Mol Biol Rep 28:176–183
Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH, Ueguchi-Tanaka M, Suzuki H, Katoh E, Luchi S, Kobayashi M, Maeda T, Matsuoka M, Yamaguchi I (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46:880–889
Peng J, Carol P, Richards DE, King KE, Cowling RJ, Murphy GP, Harberd NP (1997) The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev 11:3194–3205
Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP (1999) “Green revolution” genes encode mutant gibberellin response modulators. Nature 400:256–261
Piskurewicz U, Molina LL (2009) The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signal Behav 4:63–65
Pu L, Li Q, Fan X, Yang W, Xue Y (2008) The R2R3 MYB transcription factor GhMYB109 is required for cotton fiber development. Genetics 180:811–820
Pysh LD, Wysocka-Diller JW, Camilleri C, Bouchez D, Benfey PN (1999) The GRAS gene family in Arabidopsis: sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J 18:111–119
Qin YM, Hu CY, Pang Y, Kastaniotis AJ, Hiltunen KJ, Zhu YX (2007) Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell 19:1–13
Richards DE, King KE, Ait-ali T, Harberd NP (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Physiol Plant Mol Biol 52:67–88
Seagull RW, Giavalis S (2004) Pre- and post-anthesis application of exogenous hormones alters fiber production in Gossypium hirsutum L. cultivar Maxxa GTO. The. J Cotton Sci 8:105–111
Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX (2006) Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell 18:651–664
Silverstone AL, Ciampaglio CN, Sun TP (1998) The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 10:155–169
Sun TP (2008) Gibberellin metabolism, perception and signaling pathways in Arabidopsis. The Arabidopsis book. The American Society of Plant Biologists, Rockville
Sun TP, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Ann Rev Plant Biol 55:197–223
Taliercio EW, Boykin D (2007) Analysis of gene expression in cotton fiber initials. BMC Plant Biol 7:1–13
Tu LL, Zhang XL, Liu DQ, Jin SX, Cao JL, Zhu LF, Deng FL, Tan JF, Zhang CB (2007) Screening of reference genes for quantitative real time PCR in the process of fiber development and somatic embryo generation. Sci Rep 52:2379–2385
Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP (2004) DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol 135:1008–1019
Ueguchi-Tanaka M, Ashikari M, Nakajima M, Itoh H, Katoh E, Kobayashi M, Chow T, Hsing Y, Kitano H, Yamaguchi I, Matsuoka M (2005) Gibberellin insensitive Dwarf1 encodes a soluble receptor for gibberellin. Nature 437:693–698
Wang XQ, Ren GF, Li XM, Tu JL, Lin ZX, Zhang XL (2011) Development and evaluation of intron and insertion–deletion markers for Gossypium barbadense. Plant Mol Biol Rep. doi:10.1007/s11105-011-0369-3
Wen CK, Chang C (2002) Arabidopsis RGL1 encodes a negative regulator of gibberellin responses. Plant Cell 14:87–100
Wen W, Chen Q, Sun HY, Cui BM, Peng M (2011) Determination of collection standard of cotton ovules on different days before flowering. J Anhui Agric Sci 5:2611–2612
Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EMN, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19:1209–1220
Xiao YH, Li DM, Yin MH, Li XB, Zhang M, Wang YJ, Dong J, Zhao J, Luo M, Luo XY, Hou L, Hu L, Pei Y (2010) Gibberellin 20-oxidase promotes initiation and elongation of cotton fibers by regulating gibberellin synthesis. J Plant Physiol 167:829–837
Yang SS, Cheung F, Lee JJ, Ha M, Wei NE, Sze S, Stelly DM, Thaxton P, Triplett B, Town CD, Chen ZJ (2006) Accumulation of genome-specific transcripts, transcription factors and phytohormonal regulators during early stages of fiber cell development in allotetraploid cotton. Plant J 47:761–775
Zeng QW, Qin S, Song SQ, Zhang M, Xiao YH, Luo M, Hou L, Pei Y (2011) Molecular cloning and characterization of a cytokinin dehydrogenase gene from upland cotton (Gossypium hirsutum L.). Plant Mol Biol Rep. doi:10.1007/s11105-011-0308-3
Zhang F, Liu X, Zuo KJ, Zhang JQ, Sun XF, Tang KX (2011a) Molecular cloning and characterization of a novel Gossypium barbadense L. RAD-like gene. Plant Mol Biol Rep 29:324–333
Zhang X, Zhen JB, Li ZH, Kang DM, Yang YM (2011b) Expression profile of early responsive genes under salt stress in upland cotton (Gossypium hirsutum L.). Plant Mol Biol Rep 29:626–637
Acknowledgements
We would like to thank Prof. Kunbo Wang (Institute of Cotton Research, Chinese Academy of Agricultural Sciences) and Shaohui Li (Hainan Wild Cotton Plantation) for providing the wild diploid cotton seeds. This work was supported by the National Basic Research 973 Program of China (grant # 2004CB117307 and 2010CB126600 to M.P.) and the National Transgenic Animals & Plants Research Initiative (grant # 2009ZX08005-027B to M.P.).
Author information
Authors and Affiliations
Corresponding author
Additional information
Wei Wen and Baiming Cui contributed equally to this work.
Rights and permissions
About this article
Cite this article
Wen, W., Cui, B., Yu, X. et al. Functional Analysis of Cotton DELLA-Like Genes that are Differentially Regulated during Fiber Development. Plant Mol Biol Rep 30, 1014–1024 (2012). https://doi.org/10.1007/s11105-012-0412-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-012-0412-z