Abstract
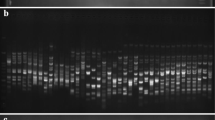
Genetic identity and relatedness of the durum wheat Graziella Ra, four Italian commercial durum cultivars (Cappelli, Grazia, Flaminio and Svevo) and Kamut were evaluated using amplified fragment length polymorphisms (AFLPs), simple sequence repeats (SSRs) and α-gliadin gene sequence analysis. Our primary objective was to study molecular genetic diversity in such a set of wheats including three modern (Grazia, Flaminio and Svevo) and three older (Cappelli, Kamut and Graziella Ra) durum accessions. Specifically, we aimed at determining the relationship between the historic accession Graziella Ra and Kamut, which is considered an ancient relative of the durum subspecies. Obtained results revealed that (1) both AFLP and SSR molecular markers detected highly congruent patterns of genetic diversity among the accessions showing nearly similar efficiency; (2) for AFLPs, percentage of polymorphic loci within accession ranged from 6.57% to 19.71% (mean, 12.77%) and for SSRs, from 0% to 57.14% (mean, 28.57%); (3) principal component analysis of genetic distance among accessions showed the first two axes accounting for 58.03% (for AFLPs) and 61.60% (for SSRs) of the total variability; (4) for AFLPs, molecular variance was partitioned into 80% (variance among accessions) and 20% (within accession) and for SSRs, into 73% (variance among accessions) and 27% (within accession); (5) cluster analysis of AFLPs and SSRs datasets displayed Graziella Ra and Kamut constantly grouped into the same cluster; and (6) molecular comparison of α-gliadin gene sequences showed Graziella Ra and Kamut in separate clusters. All these findings support the hypothesis that Graziella Ra and Kamut, although very similar, at least in the little part of the genome investigated by molecular markers employed in this study, might be considered as distinct accessions.



Similar content being viewed by others
References
Baraket G, Chatti K, Saddoud O, Abdelkarim AB, Mars M, Trifi M, Hannachi AS (2011) Comparative assessment of SSR and AFLP markers for evaluation of genetic diversity and conservation of fig, Ficus carica L., genetic resources in Tunisia. Plant Mol Biol Rep 29(1):171–184
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Boutin-Ganache I, Raposo M, Raymond M, Deschepper CF (2001) M13-tailed primers improve the readability and usability of microsatellite analyses performed with two different allele-sizing methods. Biotechniques 31:24–28
Drummond AJ, Rambaut A (2007) BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7:1214–1257
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Fukatsu E, Isoda K, Hirao T, Takahashi M, Watanabe A (2005) Development and characterization of simple sequence repeat DNA markers for Zelkova serrata. Mol Ecol Notes 5:378–380
Gregorini A, Colomba MS, Ellis HJ, Ciclitira PJ (2009) Immunogenicity characterization of two ancient wheat α-gliadin peptides related to coeliac disease. Nutrients 1:276–290
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Koning F (2005) Celiac disease: caught between a rock and a hard place. Gastroenterology 129:1294–1301
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Miller MP (1997) Tools for population genetic analyses (TFPGA 1.3): a windows program for the analysis of allozyme and molecular population genetic data. Available at http://www.marksgeneticsoftware.net/
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Peakall R, Smouse PE (2006) GenAlEx6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalsky A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Schuelke M (2000) An economic method for the fluorescent labelling of PCR fragments. Nat Biotechnol 18:233–234
Shapiro B, Rambaut A, Drummond AJ (2006) Choosing appropriate substitution models for the phylogenetic analysis of protein-coding sequences. Mol Biol Evol 22:7–9
Sharma AD, Gill PK, Singh P (2002) DNA isolation from dry and fresh samples of polysaccharide-rich plants. Plant Mol Biol Rep 20(4):415a–415f
Sneath PHA, Sokal RR (1973) Numerical taxonomy—the principles and practice of numerical classification. WH Freeman, San Francisco
Stallknecht GF, Gilbertson KM, Ranney JE (1996) Alternative wheat cereals as food grains: Einkorn, Emmer, Spelt, Kamut, and Triticale. In: Janick J (ed) Progress in new crops. ASHS, Alexandria, pp 156–170
Stephenson P, Glenn B, Kirby J, Collins A, Devosk K, Busso C, Gale M (1998) Fifty new microsatellite loci for the wheat genetic map. Theor Appl Genet 97:946–949
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. doi:10.1093/molbev/msr121. Available at http://www.kumarlab.net/publications
Von Buren M (2001) Polymorphism in two homeologous gamma-gliadin genes and the evolution of cultivated wheat. Genet Res Crop Evol 48:205–220
Vos P, Hogers R, Bleeker M, Reijans M, van der Lee TH, Hornes M, Frijters A, Pot J, Peleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23:4407–4414
Acknowledgements
We are grateful to P. Bianchi for Fig. 1. This research was supported by CIPE 20/04–DGR 438/2005 to M.S. Colomba and A. Gregorini.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Colomba, M., Vischi, M. & Gregorini, A. Molecular Characterization and Comparative Analysis of Six Durum Wheat Accessions Including Graziella Ra. Plant Mol Biol Rep 30, 168–175 (2012). https://doi.org/10.1007/s11105-011-0328-z
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11105-011-0328-z