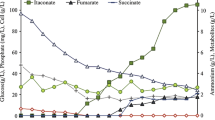
Abstract
Background and aims
The endophytic diazotrophic strain CBAmC of Nitrospirillum amazonense has been reported as a plant growth promoter of sugarcane variety RB867515 when grown under field conditions. The present work aimed to assess the influence of apoplast fluid from RB867515 on the transcriptomic and proteomic profiles of CBAmC cultured in vitro.
Methods
RNA-Seq in Ion Proton™ and ESI-LC-MS/MS peptide analysis were used to evaluate the transcriptomic and proteomic profiles, respectively, of CBAmC exposed for 2 h to the sugarcane apoplast fluid.
Results
The bacterial transcriptomic and proteomic profiles were well correlated. The overall response of CBAmC to the apoplast fluid included overexpression of defense systems against reactive oxygen species (ROS) and osmotic stress, RND efflux pumps for toxic compounds, Sec and Tat secretory systems, and assimilative metabolism of iron. In contrast, active transporters of organic compounds, chemotaxis system and flagellum structure were underexpressed.
Conclusions
The bacterial metabolic pathways / functions activated in response to the sugarcane apoplast fluid are most likely related to its adaptation to the peculiar characteristics of the fluid. The activation of some of those functions could be determinant for its adaptation to the sugarcane apoplastic niche, and perhaps be involved in the previously observed effect of promoting plant growth.








Similar content being viewed by others
Data availability
RNA-Seq data have been deposited to the NCBI’s Gene Expression Omnibus (GEO) database (Edgar et al. 2002) under the serial accession number GSE130321. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Perez-Riverol et al. 2019) partner repository with the dataset identifier PXD013518.
References
Ali S, Duan J, Charles TC, Glick BR (2014) A bioinformatics approach to the determination of genes involved in endophytic behavior in Burkholderia spp. J Theor Biol 343:193–198
Alves M, Ponce GH, Silva MA, Ensinas AV (2015) Surplus electricity production in sugarcane mills using residual bagasse and straw as fuel. Energy 91:751–757
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Bahlawane C, Baumgarth B, Serrania J, Rüberg S, Becker A (2008a) Fine-tuning of galactoglucan biosynthesis in Sinorhizobium meliloti by differential WggR (ExpG)-, PhoB-, and MucR-dependent regulation of two promoters. J Bacteriol 190:3456–3466
Bahlawane C, McIntosh M, Krol E, Becker A (2008b) Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol Plant-Microbe Interact 21:1498–1509
Baldani JI, Cruz LM, Vidal MS, Simões-Araújo JL, Schwab S, dos Teixeira, KRS, Souza EM, Pedrosa FO, Farinelli L, INCT-FBN consortium (2010) Draft genome sequence of six endophytic nitrogen-fixing bacteria with potential biofertilizer application in non-leguminous plants. In: International symposium on biological nitrogen fxation with non-legumes, 12th, Armação dos Búzios
Barnett MJ, Toman CJ, Fisher RF, Long SR (2004) A dual-genome symbiosis chip for coordinate study of signal exchange and development in a prokaryote–host interaction. Proceedings of the National Academy of Sciences USA 101:16636–16641
Becker A, Bergès H, Krol E, Bruand C, Rüberg S, Capela D, Lauber E, Meilhoc E, Ampe F, de Bruijn FJ, Fourment J, Francez-Charlot A, Kahn D, Küster H, Liebe C, Pühler A, Weidner S, Batut J (2004) Global changes in gene expression in Sinorhizobium meliloti 1021 under microoxic and symbiotic conditions. Mol Plant-Microbe Interact 17:292–303
Bertalan M, Albano R, de Pádua V, Rouws L, Rojas C, Hemerly A, Teixeira K, Schwab S, Araujo J, Oliveira A, others (2009) Complete genome sequence of the sugarcane nitrogen-fixing endophyte Gluconacetobacter diazotrophicus Pal5. BMC Genomics 10:450
Boddey RM, Urquiaga S, Alves BJR, Reis V (2003) Endophytic nitrogen fixation in sugarcane: present knowledge and future applications. Plant Soil 252:139–149
Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FCP, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M (2001) Minimum information about a microarray experiment (MIAME)—toward standards for microarray data. Nat Genet 29:365–371
Burse A, Weingart H, Ullrich MS (2004) The phytoalexin-inducible multidrug efflux pump AcrAB contributes to virulence in the fire blight pathogen, Erwinia amylovora. Mol Plant-Microbe Interact 17:43–54
Calderan-Rodrigues MJ, Jamet E, Bonassi MBCR, Guidetti-Gonzalez S, Begossi AC, Setem LV, Franceschini LM, Fonseca JG, Labate CA (2014) Cell wall proteomics of sugarcane cell suspension cultures. Proteomics 14:738–749
Camilios-Neto D, Bonato P, Wassem R, Tadra-Sfeir MZ, Brusamarello-Santos LCC, Valdameri G, Donatti L, Faoro H, Weiss VA, Chubatsu LS, Pedrosa FO, Souza EM (2014) Dual RNA-seq transcriptional analysis of wheat roots colonized by Azospirillum brasilense reveals up-regulation of nutrient acquisition and cell cycle genes. BMC Genomics 15:378
Cassán F, Vanderleyden J, Spaepen S (2014) Physiological and agronomical aspects of phytohormone production by model plant-growth-promoting rhizobacteria (PGPR) belonging to the genus Azospirillum. J Plant Growth Regul 33:440–459
Chaves VA, dos Santos SG, Schultz N, Pereira W, Sousa JS, Monteiro RC, Reis VM (2015) Initial development of two sugarcane varieties inoculated with diazotrophic bacteria. Revista Brasileira de Ciência do Solo 39:1595–1602
Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676
Cordeiro FA, Tadra-Sfeir MZ, Huergo LF, de Oliveira PF, Monteiro RA, de Souza EM (2013) Proteomic analysis of Herbaspirillum seropedicae cultivated in the presence of sugar cane extract. J Proteome Res 12:1142–1150
Coutinho BG, Licastro D, Mendonça-Previato L, Cámara M, Venturi V (2015) Plant-influenced gene expression in the rice endophyte Burkholderia kururiensis M130. Mol Plant-Microbe Interact 28:10–21
da Silva Girio LA, Ferreira Dias FL, Reis VM, Urquiaga S, Schultz N, Bolonhezi D, Mutton MA (2015) Plant growth-promoting bacteria and nitrogen fertilization effect on the initial growth of sugarcane from pre-sprouted seedlings. Pesq Agrop Brasileira 50:33–43
da Silva PRA, Simões-Araújo JL, Vidal MS, Cruz LM, de Souza EM, Baldani JI (2018a) Draft genome sequence of Paraburkholderia tropica Ppe8 strain, a sugarcane endophytic diazotrophic bacterium. Braz J Microbiol 49:210–211
da Silva PRA, Vidal MS, de Paula SC, Polese V, Tadra-Sfeir MZ, de Souza EM, Simões-Araújo JL, Baldani JI (2018b) Sugarcane apoplast fluid modulates the global transcriptional profile of the diazotrophic bacteria Paraburkholderia tropica strain Ppe8. PLoS One 13:e0207863
Davidson AL, Dassa E, Orelle C, Chen J (2008) Structure, function, and evolution of bacterial ATP-binding cassette systems. Microbiol Mol Biol Rev 72:317–364
de Souza RSC, Okura VK, Armanhi JSL, Jorrín B, Lozano N, da Silva MJ, González-Guerrero M, de Araújo LM, Verza NC, Bagheri HC, Imperial J, Arruda P (2016) Unlocking the bacterial and fungal communities assemblages of sugarcane microbiome. Sci Rep 6:28774
Delmotte N, Knief C, Chaffron S, Innerebner G, Roschitzki B, Schlapbach R, von Mering C, Vorholt JA (2009) Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. U. S. A. 106:16428–16433
diCenzo GC, Finan TM (2017) The divided bacterial genome: structure, function, and evolution. Microbiol Mol Biol Rev 81:e00019–17
Dong Z, Canny MJ, McCully ME, Roboredo MR, Cabadilla CF, Ortega E, Rodes R (1994) A nitrogen-fixing endophyte of sugarcane stems (a new role for the apoplast). Plant Physiol 105:1139–1147
dos Santos MF, de Pádua VLM, de Matos NE, Hemerly AS, Domont GB (2010) Proteome of Gluconacetobacter diazotrophicus co-cultivated with sugarcane plantlets. J Proteome 73:917–931
dos Santos SG, da Silva RF, da Fonseca CS, Pereira W, Santos LA, Reis VM (2017) Development and nitrate reductase activity of sugarcane inoculated with five diazotrophic strains. Arch Microbiol 199:863–873
Dressaire C, Moreira RN, Barahona S, Matos APA, de Arraiano CM (2015) BolA is a transcriptional switch that turns off motility and turns on biofilm development. mBio 6:e02352–14
Drogue B, Sanguin H, Borland S, Prigent-Combaret C, Wisniewski-Dyé F (2014) Genome wide profiling of Azospirillum lipoferum 4B gene expression during interaction with rice roots. FEMS Microbiol Ecol 87:543–555
Eda S, Mitsui H, Minamisawa K (2011) Involvement of the SmeAB multidrug efflux pump in resistance to plant antimicrobials and contribution to nodulation competitiveness in Sinorhizobium meliloti. Appl Environ Microbiol 77:2855–2862
Edgar R, Domrachev M, Lash AE (2002) Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210
Fan B, Carvalhais LC, Becker A, Fedoseyenko D, von Wirén N, Borriss R (2012) Transcriptomic profiling of Bacillus amyloliquefaciens FZB42 in response to maize root exudates. BMC Microbiol 12:116
Haslam RP, Downie AL, Raveton M, Gallardo K, Job D, Pallett KE, John P, Parry MAJ, Coleman JOD (2003) The assessment of enriched apoplastic extracts using proteomic approaches. Ann Appl Biol 143:81–91
James EK (2000) Nitrogen fixation in endophytic and associative symbiosis. Field Crop Res 65:197–209
James EK, Reis VM, Olivares FL, Baldani JI, Döbereiner J (1994) Infection of sugar cane by the nitrogen-fixing bacterium Acetobacter diazotrophicus. J Exp Bot 45:757–766
Kanehisa M, Goto S (2000) KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Khare E, Mishra J, Arora NK (2018) Multifaceted interactions between endophytes and plant: developments and prospects. Front Microbiol 9:2732
Kierul K, Voigt B, Albrecht D, Chen X-H, Carvalhais LC, Borriss R (2015) Influence of root exudates on the extracellular proteome of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Microbiology 161:131–147
Koebnik R (2005) TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol 13:343–347
Lee I-J, Foster KR, Morgan PW (1998) Photoperiod control of gibberellin levels and flowering in sorghum. Plant Physiol 116:1003–1011
Lery LMS, Hemerly AS, Nogueira EM, von Krüger WMA, Bisch PM (2011) Quantitative proteomic analysis of the interaction between the endophytic plant-growth-promoting bacterium Gluconacetobacter diazotrophicus and sugarcane. Mol Plant-Microbe Interact 24:562–576
Levy A, Conway JM, Dangl JL, Woyke T (2018) Elucidating bacterial gene functions in the plant microbiome. Cell Host Microbe 24:475–485
Lill R (2009) Function and biogenesis of iron–Sulphur proteins. Nature 460:831–838
Lin S-Y, Hameed A, Shen F-T, Liu Y-C, Hsu Y-H, Shahina M, Lai W-A, Young C-C (2014) Description of Niveispirillum fermenti gen. nov., sp. nov., isolated from a fermentor in Taiwan, transfer of Azospirillum irakense (1989) as Niveispirillum irakense comb. nov., and reclassification of Azospirillum amazonense (1983) as Nitrospirillum amazonense gen. nov. Antonie Van Leeuwenhoek 105:1149–1162
Lindemann A, Koch M, Pessi G, Müller AJ, Balsiger S, Hennecke H, Fischer H-M (2010) Host-specific symbiotic requirement of BdeAB, a RegR-controlled RND-type efflux system in Bradyrhizobium japonicum. FEMS Microbiol Lett 312:184–191
López-Guerrero MG, Ormeño-Orrillo E, Acosta JL, Mendoza-Vargas A, Rogel MA, Ramírez MA, Rosenblueth M, Martínez-Romero J, Martínez-Romero E (2012) Rhizobial extrachromosomal replicon variability, stability and expression in natural niches. Plasmid 68:149–158
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550
Magalhães FM, Baldani JI, Souto SM, Kuykendall JR, Döbereiner J (1983) A new acid-tolerant Azospirillum species. An Acad Bras Cienc 55:417–430
Maier T, Güell M, Serrano L (2009) Correlation of mRNA and protein in complex biological samples. FEBS Lett 583:3966–3973
Martínez-Flores I, Cano R, Bustamante VH, Calva E, Puente JL (1999) The ompB operon partially determines differential expression of OmpC in Salmonella typhi and Escherichia coli. J Bacteriol 181:556–562
McClure R, Balasubramanian D, Sun Y, Bobrovskyy M, Sumby P, Genco CA, Vanderpool CK, Tjaden B (2013) Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res 41:e140
Mendes R, Pizzirani-Kleiner AA, Araujo WL, Raaijmakers JM (2007) Diversity of cultivated endophytic bacteria from sugarcane: genetic and biochemical characterization of Burkholderia cepacia complex isolates. Appl Environ Microbiol 73:7259–7267
Morrone D, Chambers J, Lowry L, Kim G, Anterola A, Bender K, Peters RJ (2009) Gibberellin biosynthesis in bacteria: separate ent-copalyl diphosphate and ent-kaurene synthases in Bradyrhizobium japonicum. FEBS Lett 583:475–480
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5:621–628
Moutia J-FY, Saumtally S, Spaepen S, Vanderleyden J (2010) Plant growth promotion by Azospirillum sp. in sugarcane is influenced by genotype and drought stress. Plant Soil 337:233–242
Nanjo Y, Skultety L, Uváčková L, Klubicová K, Hajduch M, Komatsu S (2012) Mass spectrometry-based analysis of proteomic changes in the root tips of flooded soybean seedlings. J Proteome Res 11:372–385
Oliveira ALM, Urquiaga S, Döbereiner J, Baldani JI (2002) The effect of inoculating endophytic N2-fixing bacteria on micropropagated sugarcane plants. Plant Soil 242:205–215
Oliveira ALM, Canuto EL, Urquiaga S, Reis VM, Baldani JI (2006) Yield of micropropagated sugarcane varieties in different soil types following inoculation with diazotrophic bacteria. Plant Soil 284:23–32
Oliveira ALM, Stoffels M, Schmid M, Reis VM, Baldani JI, Hartmann A (2009) Colonization of sugarcane plantlets by mixed inoculations with diazotrophic bacteria. Eur J Soil Biol 45:106–113
Pankievicz VCS, Camilios-Neto D, Bonato P, Balsanelli E, Tadra-Sfeir M, Faoro H, Chubatsu LS, Donatti L, Wajnberg G, Passetti F, Monteiro RA, de Pedrosa FO, Souza EM (2016) RNA-seq transcriptional profiling of Herbaspirillum seropedicae colonizing wheat (Triticum aestivum) roots. Plant Mol Biol 90:589–603
Paungfoo-Lonhienne C, Lonhienne TGA, Yeoh YK, Donose BC, Webb RI, Parsons J, Liao W, Sagulenko E, Lakshmanan P, Hugenholtz P, Schmidt S, Ragan MA (2016) Crosstalk between sugarcane and a plant-growth promoting Burkholderia species. Sci Rep 6:37389
Pedula RO, Schultz N, Monteiro RC, Pereira W, de Araujo AP, Urquiaga S, Reis VM (2016) Growth analysis of sugarcane inoculated with diazotrophic bacteria and nitrogen fertilization. Afr J Agric Res 11:2786–2795
Pereira W, Leite JM, de Hipólito GS, dos Santos CLR, Reis VM (2013) Biomass accumulation in sugarcane varieties inoculated with different strains of diazotrophs. Rev Ciênc Agron 44:363–370
Perez-Riverol Y, Csordas A, Bai J, Bernal-Llinares M, Hewapathirana S, Kundu DJ, Inuganti A, Griss J, Mayer G, Eisenacher M, Pérez E, Uszkoreit J, Pfeuffer J, Sachsenberg T, Yılmaz Ş, Tiwary S, Cox J, Audain E, Walzer M, Jarnuczak AF, Ternent T, Brazma A, Vizcaíno JA (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res 47:D442–D450
Postle K (2007) TonB system, in vivo assays and characterization. Methods Enzymol 422:245–269
Ramachandran VK, East AK, Karunakaran R, Downie JA, Poole PS (2011) Adaptation of Rhizobium leguminosarum to pea, alfalfa and sugar beet rhizospheres investigated by comparative transcriptomics. Genome Biol 12:R106
Reinhold-Hurek B, Hurek T (2011) Living inside plants: bacterial endophytes. Curr Opin Plant Biol 14:435–443
Reis VM, Olivares FL, Döbereiner J (1994) Improved methodology for isolation of Acetobacter diazotrophicus and confirmation of its endophytic habitat. World J Microbiol Biotechnol 10:401–405
Reis VM, Baldani JI, Urquiaga S (2009) Recomendação de uma mistura de estirpes de cinco bactérias fixadoras de nitrogênio para inoculação de cana-de-açúcar: Gluconacetobacter diazotrophicus (BR 11281), Herbaspirillum seropedicae (BR 11335), Herbaspirillum rubrisubalbicans (BR 11504), Azospirillum amazonense (BR 11145) e Burkholderia tropica (BR 11366). Circular Técnica - Embrapa Agrobiologia 30:1–4
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Rodrigues EP, Rodrigues LS, Oliveira ALM, Baldani VLD, dos Teixeira KRS, Urquiaga S, Reis VM (2008) Azospirillum amazonense inoculation: effects on growth, yield and N2 fixation of rice (Oryza sativa L.). Plant Soil 302:249–261
Rosconi F, Souza EM, Pedrosa FO, Platero RA, González C, González M, Batista S, Gill PR, Fabiano ER (2006) Iron depletion affects nitrogenase activity and expression of nifH and nifA genes in Herbaspirillum seropedicae. FEMS Microbiol Lett 258:214–219
Rosenblueth M, Martínez-Romero E (2006) Bacterial endophytes and their interactions with hosts. Mol Plant-Microbe Interact 19:827–837
Rüberg S, Pühler A, Becker A (1999) Biosynthesis of the exopolysaccharide galactoglucan in Sinorhizobium meliloti is subject to a complex control by the phosphate-dependent regulator PhoB and the proteins ExpG and MucR. Microbiology 145(Pt 3):603–611
Salah Ud-Din AIM, Roujeinikova A (2017) Methyl-accepting chemotaxis proteins: a core sensing element in prokaryotes and archaea. Cell Mol Life Sci 74:3293–3303
Sattelmacher B, Horst WJ (2007) The apoplast of higher plants: compartment of storage, transport and reactions. Springer, Dordrecht
Schultz N, de Morais RF, da Silva JA, Baptista RB, Oliveira RP, Leite JM, Pereira W, de Carneiro Júnior JB, BJR A, Baldani JI, Boddey RM, Urquiaga S, Reis VM (2012) Avaliação agronômica de variedades de cana-de-açúcar inoculadas com bactérias diazotróficas e adubadas com nitrogênio. Pesq Agrop Brasileira 47:261–268
Schultz N, da Silva JA, Sousa JS, Monteiro RC, Oliveira RP, Chaves VA, Pereira W, da Silva MF, Baldani JI, Boddey RM, Reis VM, Urquiaga S (2014) Inoculation of sugarcane with diazotrophic bacteria. Revista Brasileira de Ciência do Solo 38:407–414
Schultz N, Pereira W, Reis VM, Urquiaga SS (2016) Produtividade e diluição isotópica de 15N em cana-de-açúcar inoculada com bactérias diazotróficas. Pesq Agrop Brasileira 51:1594–1601
Schwab S, Terra LA, Baldani JI (2018) Genomic characterization of Nitrospirillum amazonense strain CBAmC, a nitrogen-fixing bacterium isolated from surface-sterilized sugarcane stems. Mol Gen Genomics 293:997–1016
Shidore T, Dinse T, Öhrlein J, Becker A, Reinhold-Hurek B (2012) Transcriptomic analysis of responses to exudates reveal genes required for rhizosphere competence of the endophyte Azoarcus sp. strain BH72. Environ Microbiol 14:2775–2787
Shimizu K (2013) Regulation systems of bacteria such as Escherichia coli in response to nutrient limitation and environmental stresses. Metabolites 4:1–35
Soares C de P, Rodrigues EP, de Paula Ferreira J, Araújo JLS, Rouws LFM, Baldani JI, Vidal MS (2015) Tn5 insertion in the tonB gene promoter affects iron-related phenotypes and increases extracellular siderophore levels in Gluconacetobacter diazotrophicus. Arch Microbiol 197:223–233
Spaepen S, Vanderleyden J, Remans R (2007) Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol Rev 31:425–448
Stoitsova SO, Braun Y, Ullrich MS, Weingart H (2008) Characterization of the RND-type multidrug efflux pump MexAB-OprM of the plant pathogen Pseudomonas syringae. Appl Environ Microbiol 74:3387–3393
Tadra-Sfeir MZ, Faoro H, Camilios-Neto D, Brusamarello-Santos L, Balsanelli E, Weiss V, Baura VA, Wassem R, Cruz LM, De Oliveira PF, Souza EM, Monteiro RA (2015) Genome wide transcriptional profiling of Herbaspirillum seropedicae SmR1 grown in the presence of naringenin. Front Microbiol 6:491
Takeshima K, Hidaka T, Wei M, Yokoyama T, Minamisawa K, Mitsui H, Itakura M, Kaneko T, Tabata S, Saeki K, Oomori H, Tajima S, Uchiumi T, Abe M, Tokuji Y, Ohwada T (2013) Involvement of a novel genistein-inducible multidrug efflux pump of Bradyrhizobium japonicum early in the interaction with Glycine max (L.) Merr. Microbes Environ 28:414–421
Taylor CF, Paton NW, Lilley KS, Binz P-A, Julian RK Jr, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, Dunn MJ, Heck AJR, Leitner A, Macht M, Mann M, Martens L, Neubert TA, Patterson SD, Ping P, Seymour SL, Souda P, Tsugita A, Vandekerckhove J, Vondriska TM, Whitelegge JP, Wilkins MR, Xenarios I, Yates JR III, Hermjakob H (2007) The minimum information about a proteomics experiment (MIAPE). Nat Biotechnol 25:887–893
Tejera N, Ortega E, Rodes R, Lluch C (2006) Nitrogen compounds in the apoplastic sap of sugarcane stem: some implications in the association with endophytes. J Plant Physiol 163:80–85
Urquiaga S, Cruz KH, Boddey RM (1992) Contribution of nitrogen fixation to sugar cane: nitrogen-15 and nitrogen-balance estimates. Soil Sci Soc Am J 56:105–114
Urquiaga S, Xavier RP, de Morais RF, Batista RB, Schultz N, Leite JM, Maia e Sá J, Barbosa KP, de Resende AS, BJR A, Boddey RM (2012) Evidence from field nitrogen balance and 15N natural abundance data for the contribution of biological N2 fixation to Brazilian sugarcane varieties. Plant Soil 356:5–21
Vessey JK (2003) Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255:571–586
Vogel C, Marcotte EM (2012) Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet 13:227–232
von Kovats E (1958) Gas-chromatographische charakterisierung organischer verbindungen. Teil 1: retentionsindices aliphatischer halogenide, alkohole, aldehyde und ketone. Helvetica Chimica Acta 41:1915–1932
Wade JT, Grainger DC (2014) Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol 12:647–653
Welbaum GE, Meinzer FC (1990) Compartmentation of solutes and water in developing sugarcane stalk tissue. Plant Physiol 93:1147–1153
Wexler M, Yeoman KH, Stevens JB, Luca NGD, Sawers G, Johnston AWB (2001) The Rhizobium leguminosarum tonB gene is required for the uptake of siderophore and haem as sources of iron. Mol Microbiol 41:801–816
Xu Q, Dziejman M, Mekalanos JJ (2003) Determination of the transcriptome of Vibrio cholerae during intraintestinal growth and midexponential phase in vitro. Proc Natl Acad Sci U S A 100:1286–1291
Yoder-Himes DR, Konstantinidis KT, Tiedje JM (2010) Identification of potential therapeutic targets for Burkholderia cenocepacia by comparative transcriptomics. PLoS One 5:e8724
Acknowledgements
The authors thank Alexandre C. Baraúna for helping in the apoplast fluid preparation. This study was partially supported by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES-L.A.T. doctorate fellowship, call CAPES/Embrapa no. 015/2014), and Brazilian Agricultural Research Corporation (Embrapa) project code 02.13.08.006.00.00.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Jerri Edson Zilli
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Terra, L.A., de Soares, C.P., Meneses, C.H.S.G. et al. Transcriptome and proteome profiles of the diazotroph Nitrospirillum amazonense strain CBAmC in response to the sugarcane apoplast fluid. Plant Soil 451, 145–168 (2020). https://doi.org/10.1007/s11104-019-04201-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04201-y