Abstract
Aims
Tree roots contribute large quantities of biomass in forests and are important drivers of different ecosystem processes. However, estimation of root biomass remains a challenge. We have developed a method to quantify fine root biomass of trees, tested on Pinus sylvestris L. and Betula sp. in soils in natural environment, by applying specific qPCR primers to soil DNA extracts.
Methods
We first validated the molecular probes in the laboratory by constructing regression equations among known root biomass in soil and the number of gene copies obtained by qPCR. Then, to test the applicability of the method in an ecosystem setting and because Pinus and Betula are important components of boreal forests after wildfire, we quantified fine root biomass from these trees in soils from forests with different degrees of tree mortality after wildfire.
Results
The new primers specifically amplified ITS (Internal Transcribed Spacer) markers from DNA extracted from fine roots of Pinus and Betula, and regression equations allowed us to calculate root biomass in soil from the number of gene copies obtained by qPCR. Root biomass of Pinus correlated positively with fire-induced canopy mortality, confirming the adequacy of the method. The biomass of Betula roots in soil did not differ across the fire severity gradient due to an overall low and stochastic presence of birch, and did not relate to the establishment of new birch seedlings.
Conclusions
This study shows that DNA-based qPCR methods can be used for rapid, quantitative and species-specific analysis of tree root biomass in natural forest ecosystems.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Tree roots are major contributors to the total biomass of forests, and an important fraction of carbon (C) assimilated by plants through photosynthesis is relocated to roots and their symbionts (McCormack et al. 2015). For example, in boreal forest, C accumulating below ground is primarily originating from root-derived sources (Clemmensen et al. 2013; Kyaschenko et al. 2018). Although the production and biomass of tree roots are important drivers of various ecosystem processes, quantitative knowledge about roots is much more limited than for aboveground constituents of trees (Yuan and Chen 2010). This lack of information can partly be ascribed to methodological challenges in estimation of root biomass and production (Makkonen and Helmisaari 1999).
There is an array of different methods to study roots, but no consensus on how to best estimate root biomass and production (Addo-Danso et al. 2016). Most of the traditional methods, such as soil cores, rhizotrons and ingrowth-cores, are time consuming and limited by low accuracy and precision (Haling et al. 2011). In the last decade, DNA-based techniques have been implemented to identify roots of various plant species (Jones et al. 2011), and real-time PCR (qPCR) has been used to assess species diversity of roots in mixed samples (Mommer et al. 2008; Randall et al. 2014; Zeng et al. 2015). However, because root extraction is labour intensive, Riley et al. (2010) proposed a method to quantify roots directly in soil samples through assessing DNA extracted from soil, using specific qPCR probes. Quantification of roots by qPCR has previously been applied in agricultural soils in greenhouse experiments (Riley et al. 2010; Haling et al. 2011), and there is to date no understanding of how applicable this method is for assessing root biomass on an area basis in natural ecosystems.
Boreal forests are formed by a relatively low number of tree species. In Fennoscandia, Scots pine (Pinus sylvestris L.) and Norway spruce (Picea abies L.) Karst.) represent the dominant conifers, while deciduous tree species within the genera Betula, Populus and Alnus typically co-occur with conifers after disturbances, such as windstorms, clear cuts or fires (Hari and Kulmala 2009). In this study, we designed and applied specific primers to amplify DNA from two common tree species (Pinus sylvestris and Betula sp) growing in undisturbed boreal forest soils and soils subjected to fire disturbances. The aim was to apply qPCR for root biomass quantification directly in forest soil samples, and enable estimations of fine root biomass on an area basis of forest ecosystems. We first validated the molecular probes in the laboratory by constructing regression equations that allowed calculating root biomass in soil from the number of gene copies obtained by qPCR. Secondly, to test the applicability of the method, we quantified tree root biomass in soils from forests recently disturbed by wildfire to varying degree, e.g. along a gradient of forest stands that differed in fire severity and canopy mortality. We evaluated how well belowground density of pine DNA reflected aboveground mortality of pine trees after fire and related our fine root biomass estimates to previous reports of direct observations from similar ecosystems. Moreover, we tested how the density of birch DNA reflected the establishment of post-fire colonizing deciduous tree seedlings. The ultimate aim of this work was to contribute to a better understanding of how suitable extraction of tree DNA directly from forest soils is for determining the relationship between species-specific tree root biomass and the presence of these species within the ecosystem.
Material and Method
Design and validation of specific primers for Pinus and Betula
Specific primers for Pinus sylvestris were designed to amplify a fragment of the ITS region of the rRNA encoding operon (Table 1). Multiple alignments of ITS sequences from different Pinaceae and Betulaceae species (obtained from the NCBI database) was conducted (MultAlin software) and different conserved regions were selected as targets for primers Fig. S1 (Online Resource). The primers were validated in “Multiprimer” (www.thermoscientificbiomultiprimer), checking for dimer formation and acceptable melting temperatures. To assess specificity, the oligos were tested in silico by blasting against the NCBI GenBank database.
Further specificity validation and determination of ITS copies to biomass conversion factors were carried out as follows. In September 2017, fine roots of different P. sylvestris trees (<2 mm in diameter) from a monodominant stand and 10 soil cores (3 cm in diameter) from a Picea abies (also within Pinaceae) dominated forest were collected near Uppsala, Sweden. In May 2018, fine roots of different Betula sp. trees and 10 soil cores under Alnus glutinosa (also within Betulaceae) were also collected. Fine roots were collected by tracking main roots attached to the trees. Additionally, as an extra negative control for the specificity tests, replicated soil samples from a wheat field (free of tree roots) were sampled (Hamnér et al. 2017). The roots were gently washed with tap water to remove soil particles. Roots and soil samples were kept at −20 °C and then freeze-dried and ball milled. Weighted aliquots of Pinus roots were mixed into P. abies soil (as a pine-free background to verify specificity), and Betula root aliquots were mixed into A. glutinosa soil (as a birch-free background) (Table S1, online resource). Different amount of roots were mixed with soil to prepare standards for root quantification in soil samples, with dry weight root/soil ratios ranging from 1:1000 to 1:10 (Table S1).
To obtain ITS amplicons for constructing plasmid standard curves, genomic DNA from 250 mg of Pinus or Betula roots was extracted by the NucleoSpin Soil Genomic DNA extraction kit (Macherey-Nagel, Düren, Germany). PCR amplification of ITS markers was conducted in 50 μl reactions containing 2.5 ng of genomic DNA, polymerase buffer (ThermoFisher), 0.75 mM MgCl2, 0.2 mM of dNTPs, 0.4 μM of forward and reverse primers (Table 1) and 0.025 U μl−1 of DreamTaq polymerase enzyme (ThermoFisher). The PCR conditions were as follows: 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 58 °C for 30 s and 72 °C for 30 s, with a final step of 72 °C for 7 min. To construct standards for qPCR quantification, purified PCR products from Pinus and Betula roots were cloned into chemically competent Escherichia coli One-Shot® TOP10, using TOPO TA cloning® (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. The plasmids were serially diluted to construct qPCR standard curves.
Real-time PCRs were performed in a Biorad iQ5 real-time PCR detector system (Bio-Rad, Richmond, CA, USA) in a total reaction volume of 20 μl. In the case of Pinus, the reaction contained EvaGreen qPCR Master Mix (Bio-Rad, USA); 250 nM of ITSp_F primer, 500 nM of ITSp_R primer, and 5 pg of DNA template. In the case of Betula, the mixture contained EvaGreen qPCR Master Mix (Bio-Rad, USA); 750 nM of ITSb_F primer, 250 nM of ITSb_R primer and 5 pg of DNA template. Primers and annealing temperatures were optimised for each tree species based on 9 combinations of three different primers concentrations in temperature gradients. The optimized amplification conditions for pine roots were: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and 56 °C for 34 s; and for birch roots: 95 °C for 30 s, followed by 40 cycles of 95 °C for 5 s, and 55 °C for 34 s. Negative controls were included.
Genomic DNA was extracted from 2 ml of the Picea or Alnus soils (as negative controls) as well as from the soil-roots mixtures, following the extraction protocol of Clemmensen et al. (2016). DNA was subjected to qPCR following the optimized conditions used for roots. Three technical replicates were run for each dilution of the DNA plasmid standard curve and for each root-soil sample. Standard curves of root mass (mg) versus ITS copy numbers were established (Fig. 1). Inhibition tests of reactions containing soil-derived DNA were performed by adding a known amount of plasmid DNA to each sample or to blanks containing water and amplifying with M13 primers (targeting primer sites in the plasmid) following the same PCR conditions as above. Inhibition was considered significant when the Ct value (the cycle threshold defined as the cycle number at which the log-linear phase could be distinguished from the background) of a sample spiked with plasmid was >0.5 log units higher than that of plasmid in water.
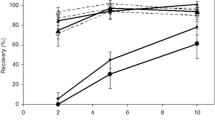
Linear relation (mean ± S.E.) between the known mass of (a) Pinus sylvestris roots and (b) Betula sp. roots in soil (mg) and the number of ITS copies obtained by qPCR in triplicates. Different amounts of roots were mixed with soil to obtain these standards for root quantification in soil samples based on soil DNA extraction (Table S1)
Study area and soil sampling
To test the applicability of the method for forest soils, we quantified tree root biomass in soil collected from each of 32 pine dominated forest stands in the Västmanland county, South-Central Sweden (N 59° 53′44″, E 16° 11′27″), representing a gradient in fire severity and mortality in the tree canopy.
Twenty-five of the stands were affected by a fire in 2014, while 7 of the stands escaped the fire. Prior to the fire all sites were dominated by P. sylvestris with scattered P. abies and Betula sp., and an understory vegetation dominated by ericaceous dwarf shrubs (Vaccinium vitis-idaea L., V. uliginosum L. and Calluna vulgaris (L.) Hull) and mosses (mainly Pleurozium schreberi (B.) Mitt.). All forest sites had undergone first-thinning operations after clearcutting (age between 40 and 60 years), are located on similar topography and are situated within an area covering approximately 78 km2, separated from each other by a minimum distance of 190 m. The soil is classified as a spodosol developed in glacial granitic till.
In May 2016, we collected 25 soil cores (3 cm in diameter) from the organic horizon in each stand. Cores were collected at one-meter intervals within a 20 × 20 m grid of each site (800 soil cores in total). Directly after sampling, all green plant parts and mineral soil were removed from the core by hand or using a knife and the entire depth of the organic horizon was retained. Soil cores were pooled, resulting in one composite sample per stand (n = 32), and kept at −20 °C until processed (mineral soils were also pooled but not used further in this study). Frozen samples were homogenized, freeze-dried and ball-milled. DNA was extracted from the finely powdered soil, and three replicates of each extract were amplified by qPCR following the procedures explained above. The root mass concentration of Pinus and Betula was estimated using the equations presented in Fig. 1a and Fig. 1b, respectively, and averaged estimates were further converted to an area basis (g of roots m−2), accounting for the total sampling area and dry mass of the 25 soil cores. The experimental procedures are presented in Fig. S2 (Online Resource).
In order to test the relationship between tree root biomass and aboveground presence of trees, a 10 m radius plot was established in the centre of each of the burnt stands. Within this plot, the proportion of fire induced mortality of pine and birch trees was recorded (percentage of alive trees from the total number of trees). In addition, the presence of pine and birch seedlings was scored as the frequency (presence/absence) in 10 × 10 cm subplots in a grid of 0.25 m2 randomly located within the larger radius plot, with seedling cover expressed as the average frequency of 8 assessed grids within each centre plot. The proportion of adult birch trees and the cover of pine seedling were not used in further analyses because of low numbers (data not shown). Additionally, we recorded the presence of post-fire vegetation. We used Pearson correlation coefficient analysis to test for the correlation between tree root biomass of Pinus and the survival of tree canopies and between tree root biomass of Betula and the frequency of new birch seedlings after wildfire.
Results
Validation of specific primers for Pinus and Betula
Standard curves were generated from plasmids containing ITS PCR products – Pinus: R2 = 0.995, PCR efficiency = 98.1%; Betula: R2 = 0.999, PCR efficiency = 101.2%.
To check for genus-level specificity of the qPCR assay, lack of amplification was confirmed in soil samples from under Picea (as a negative control for Pinus) or from soil under Alnus trees (as a negative control for Betula). Soil samples free from roots of the target species showed similar Ct values as water blanks, and significantly higher Ct values (i.e. lower starting quantity) than the mixtures of soil with the lowest amount of roots (1/1000 roots to soil) (Pinus t test: p < 0.001; Betula t test: p = 0.009).
We confirmed a positive linear relationship (R2 = 0.991 for Pinus; R2 = 0.999 for Betula) between the ITS copy numbers obtained by qPCR and the mg of roots in extracted soil mixed with known amounts of roots (Fig. 1a, b). Regression equations allowed us to calculate root mass (mg of roots) from the number of ITS copies obtained by qPCR that was, in turn, inferred from Ct values.
Field study
The two pairs of primers successfully amplified Pinus and Betula root DNA from all soil samples in each of the 32 stands without significant PCR inhibition. The average pine root biomass in the unburned plots, representative of grown forests, was 130 ± 44 g m−2 and varied from 0 to 19 g m−2 in the burned plots (Fig. 2), depending on tree mortality (Fig. 3). Estimated pine root biomass positively correlated with the survival of pine canopies (Pearson correlation: r = 0.851; P < 0.001). Root biomass of birch was 17 ± 3 g m−2 in the unburned plots (Fig. 2), and varied from 4 to 39 g of roots per m−2 in burned plots with no statistically significant correlation with the frequency of new birch seedlings (Pearson correlation: r = 0.087; P = 0.686) (Figs. 2 and 3).
Fine root mass of Pinus sylvestris (black) and Betula sp. (grey) estimated by qPCR in soils from 7 unburned plots (reference plots) and 25 burned plots. Values are means ± SE. Silhouettes have been downloaded from http://www.phylopic.org and http://www.urltarget.com
Fine root mass estimated by qPCR (g roots m−2) of (a) Pinus sylvestris and (b) Betula sp. from soils of 25 plots differing in the fire severity damage (black bars). Pinus sylvestris canopy mortality in (a) is represented by grey bars and Betula sp. seedling cover in (b) is represented by white bars. Silhouettes have been downloaded from http://www.phylopic.org and http://www.urltarget.com
Discussion
Our work is the first to demonstrate that DNA-based qPCR methods can be used for rapid, quantitative and species specific analysis of tree root biomass on an areal basis directly in soil samples of natural environments. We applied the method in artificial samples and field samples of soils from a fire severity gradient, demonstrating the potential use of specific primers to investigate fine root densities of selected tree species in forest soils. The method may be particularly useful in holistic studies of soil ecology, in which microbial communities and root biomass can be assessed and compared from the same DNA extracts.
Molecular techniques have previously been used for quantification of root biomass of mixed plant species in greenhouse experiments and agricultural soils, and advantages over other techniques have been discussed (Riley et al. 2010; Haling et al. 2011). Moreover, these authors found that the DNA detected by the qPCR assays primarily originated from living cells, which overcomes the major difficulty of determining living versus dead root mass by morphological characteristics. Other studies have demonstrated that qPCR can be used to estimate the relative proportion of a given species in mixed fine-root samples from (sub) tropical forest (Zeng et al. 2017) and greenhouse experiments (Mommer et al. 2008). Our study is the first to utilize qPCR to quantify fine root mass in soils from natural environments, such as forest soils. Haling et al. (2011) stated that the method might be more applicable to mono-dominant communities or communities with low numbers of plant species and less suitable in highly diverse plant communities. In our experiment, we focused on Pinus and Betula, e.g. the tree species that survived the large wildfire, and that are also typical early successional tree species following fire. The primers designed and validated in this study successfully amplified roots of Pinus and Betula from soils of forests that inhabited plant roots of several other plant genus, e.g. species of graminoids, dwarf shrubs, and forbs. The stable copy number versus root biomass relationships found across different soil samples in our study suggests that assay calibrations only needs to be done once per primer pair. Further, several pairs of primers can be used simultaneously on the same DNA extract if several plant species are of interest. With its high interspecific variation, the ITS region offers great opportunities to easily design specific primers for a multitude of species, and with some more efforts, broader primers that target higher taxonomic levels can also be designed.
After evaluating standardized soil-root mixtures for each of the two tree species (Pinus and Betula), in the laboratory with satisfactory results, we applied the qPCR method to quantify the root biomass of Pinus and Betula in soil samples collected from forest stands experiencing differing burn severity. The average root biomass found in the unburned reference plots (130 g m−2) was within the range earlier estimated by root sampling. According to Finér et al. (2007) the pine fine-root biomass (<2 mm in diameter) in a variety of boreal forests ranged between 26 and 467 g m−2 with an average of 229 g m−2. Makkonen and Helmisaari (1999) reported a three-year average of P. sylvestris fine-root biomass of 261 g m−2 and 241 g m−2 by measuring soil cores and ingrowth cores, respectively. In 15–20 and 120-years old P. sylvestris forests, fine-root biomass of 26 g m−2 and 123 g m−2, respectively, were reported (Persson 1983). However, further validation including different sampling methods like rhizotrons and/or ingrowth-cores would increase the precision of the estimated root production, as the optimal sampling technique may differ among species, e.g. due to differences in root length (Kalliokoski et al. 2008). Still, root biomass assessments are variable among different methods of assessment, and it remains a challenge to accurately determine belowground production in forests (Addo-Danso et al. 2016). Fire significantly reduced the root biomass of Pinus in line with canopy mortality, and the death of pine roots could thus be a good indicator of fire severity and an important variable when investigating fire responses of the belowground system. When canopy mortality exceeded 50%, little root DNA was detected, presumably due to the fact that surviving trees had severely reduced foliage. This suggests that root DNA is a fast indicator of responses of living root to disturbances such as wildfire. Further, ectomycorrhizal trees frequently display positive plant-soil feedback (Bennett et al. 2017), implying that living roots may facilitate establishment of conspecific seedlings. Thus, plant succession after varying burn severity could potentially be different depending on root survival. The fire did not reduce birch root biomass compared to the unburnt stands and we were unable to detect any relationship between fine root biomass of birch and burn severity or seedling density. The lack of correlation was probably due to the fact that we sampled shortly after fire (e.g. two years after the fire,) when the density and biomass of birch seedlings were still at a low level. Other possibilities that cannot be ruled out is amplification of birch DNA from pollen (Riley et al. 2010) or root survival of scattered adult birch trees (the mean number of alive birch trees per 10 m radius plot was 0.4 ± 0.8 for burned stands and 1.5 ± 3.7 for unburned stands).
In this work, we demonstrated the use of specific primers to quantitatively investigate root densities of Pinus and Betula in forest soils after wildfire. The low tree species richness of boreal forest makes it an ideal system to test and develop the method. We infer that the method would be suitable for estimating species-specific root biomass of trees of more diverse ecosystems as well. However, to firmly value the general suitability and use of this method, we encourage further tests under a greater variety of conditions and range of forest types, and particularly additional specificity tests for communities of closely related species. This would in turn also allow a new set of research questions being answered, that so far have been restricted by the tedious task of manual species-specific sorting of roots. Additionally, these data could optimally be obtained and used together with other DNA based soil community investigations.
References
Addo-Danso SD, Prescott CE, Smith AR (2016) Methods for estimating root biomass and production in forest and woodland ecosystem carbon studies: a review. For Ecol Manag 359:332–351. https://doi.org/10.1016/j.foreco.2015.08.015
Bennett JA, Maherali H, Reinhart KO et al (2017) Plant-soil feedbacks and mycorrhizal type influence temperate forest population dynamics. Science (80- ) 184:181–184. https://doi.org/10.1126/science.aai8212
Clemmensen KE, Bahr A, Ovaskainen O, Dahlberg A, Ekblad A, Wallander H, Stenlid J, Finlay RD, Wardle DA, Lindahl BD (2013) Roots and associated fungi drive long-term carbon sequestration in boreal Forest. Science (80- ) 339:1615–1618. https://doi.org/10.1126/science.1231923
Clemmensen KE, Ihrmark K, Mikael Brandström Durling BDL (2016) Sample Preparation for Fungal Community Analysis by High-Throughput Sequencing of Barcode Amplicons. In: Humana Press NY (ed) Microbial environmental genomics (MEG). pp 61–88
Finér L, Helmisaari HS, Lõhmus K, Majdi H, Brunner I, Børja I, Eldhuset T, Godbold D, Grebenc T, Konôpka B, Kraigher H, Möttönen MR, Ohashi M, Oleksyn J, Ostonen I, Uri V, Vanguelova E (2007) Variation in fine root biomass of three European tree species: beech (Fagus sylvatica L.), Norway spruce (Picea abies L. Karst.), and scots pine (Pinus sylvestris L.). Plant Biosyst 141:394–405. https://doi.org/10.1080/11263500701625897
Haling RE, Simpson RJ, McKay AC et al (2011) Direct measurement of roots in soil for single and mixed species using a quantitative DNA-based method. Plant Soil 348:123–137. https://doi.org/10.1007/s11104-011-0846-3
Hamnér K, Weih M, Eriksson J, Kirchmann H (2017) Field crops research in fl uence of nitrogen supply on macro- and micronutrient accumulation during growth of winter wheat. F Crop Res 213:118–129. https://doi.org/10.1016/j.fcr.2017.08.002
Hari P, Kulmala L (2009) Boreal forests and climate change. Springer, Dordrecht
Jones FA, Erickson DL, Bernal MA, et al (2011) The Roots of Diversity : Below Ground Species Richness and Rooting Distributions in a Tropical Forest Revealed by DNA Barcodes and Inverse Modeling. 6. https://doi.org/10.1371/journal.pone.0024506
Kalliokoski T, Nygren P, Sievänen R (2008) Coarse root architecture of three boreal tree species growing in mixed stands. Silva Fenn 42:189–210. https://doi.org/10.14214/sf.252
Kyaschenko J, Ovaskainen O, Ekblad A, Hagenbo A, Karltun E, Clemmensen KE, Lindahl BD (2018) Soil fertility in boreal forest relates to root-driven nitrogen retention and carbon sequestration in the mor layer. New Phytol 221:1492–1502. https://doi.org/10.1111/nph.15454
Makkonen K, Helmisaari HS (1999) Assessing fine-root biomass and production in a scots pine stand - comparison of soil core and root ingrowth core methods. Plant Soil 210:43–50. https://doi.org/10.1023/A:1004629212604
McCormack ML, Dickie IA, Eissenstat DM et al (2015) Redefining fine roots improves understanding of below-ground contributions to terrestrial biosphere processes. New Phytol 207:505–518. https://doi.org/10.1111/nph.13363
Mommer L, Wagemaker CAM, De Kroon H, Ouborg NJ (2008) Unravelling below-ground plant distributions: a real-time polymerase chain reaction method for quantifying species proportions in mixed root samples. Mol Ecol Resour 8:947–953. https://doi.org/10.1111/j.1755-0998.2008.02130.x
Persson HÅ (1983) The distribution and productivity of fine roots in boreal forests. In: Tree root systems and their mycorrhizas. Springer D (ed), pp 87–101
Randall MJ, Karst J, Pec GJ, Davis CS, Hall JC, Cahill JF Jr (2014) A molecular identification protocol for roots of boreal Forest tree species. Appl Plant Sci 2:1400069. https://doi.org/10.3732/apps.1400069
Riley IT, Wiebkin S, Hartley D, McKay AC (2010) Quantification of roots and seeds in soil with real-time PCR. Plant Soil 331:151–163. https://doi.org/10.1007/s11104-009-0241-5
Yuan ZY, Chen H (2010) Fine root biomass, production, turnover rates, and nutrient contents in boreal forest ecosystems in relation to species, climate, fertility, and stand age: literature review and meta-analyses. CRC Crit Rev Plant Sci 29:204–221. https://doi.org/10.1080/07352689.2010.483579
Zeng W, Zhou B, Lei P, Zeng Y, Liu Y, Liu C, Xiang W (2015) A molecular method to identify species of fine roots and to predict the proportion of a species in mixed samples in subtropical forests. Front Plant Sci 6:1–10. https://doi.org/10.3389/fpls.2015.00313
Zeng W, Xiang W, Zhou B, Lei P, Zeng Y (2017) Measurement of belowground diversity of fine roots in subtropical forests based on a quantitative real-time PCR (qPCR) method. Plant Soil 420:539–552. https://doi.org/10.1007/s11104-017-3402-y
Acknowledgements
This research was supported by the Swedish Research Council FORMAS (2014-01858). LPI held a post-doctoral fellowship funded by Swedish University of Agricultural Sciences.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(PDF 508 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Pérez-Izquierdo, L., Clemmensen, K.E., Strengbom, J. et al. Quantification of tree fine roots by real-time PCR. Plant Soil 440, 593–600 (2019). https://doi.org/10.1007/s11104-019-04096-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-019-04096-9