Abstract
Aims
Grasslands dominated by non-native (exotic) species have replaced purely native-dominated areas in many parts of the world forming ‘novel’ ecosystems. Altered precipitation patterns are predicted to exacerbate this trend. It is still poorly understood how soil microbial communities and their functions differ between high diversity native- and low diversity exotic-dominated sites and how altered precipitation will impact this difference.
Methods
We sampled 64 experimental grassland plots in central Texas with plant species mixtures of either all native or all exotic species; half with summer irrigation. We tested how native vs. exotic plant species mixtures and summer irrigation affected bacterial and fungal community composition and structure, the influence of niche vs. neutral processes for microbial phylotype co-occurrence (C-score analysis), and rates of phosphorus and nitrogen mineralization across an 8-year experiment.
Results
Native and exotic-dominated plots had significantly different fungal community composition and structure, but not diversity, throughout the length of the study, while changes in bacterial communities were limited to certain wet and cool years. Nitrogen and phosphorus mineralization rates were higher under native plant mixtures and correlated with the abundance of particular fungal species. Microbial communities were more structured in exotic than native grassland plots, especially for the fungal community.
Conclusions
The results indicate that conversion of native to exotic C4 dominated grasslands will more strongly impact fungal than bacterial community structure. Furthermore, these impacts can alter ecosystem functioning belowground via changes in nitrogen and phosphorus cycling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The introduction and continued spread of exotic plant species are shaping landscapes of North America leading to the establishment of ‘novel’ ecosystems, sites dominated by non-native plant species from different regions (Hobbs et al. 2006, 2009, 2013). Novel ecosystems can function differently than the native systems that they replace, and a better understanding of how native and novel ecosystems differ will help management and policy efforts (Seastedt et al. 2008; Wilsey et al. 2009, 2017). Climate patterns have become novel as well (Allan and Soden 2008), both in the timing and intensity of rainfall events, and altered rainfall is predicted to favor exotic plant species (Dukes and Mooney 1999). Climate change could have indirect or direct effects on microbial communities, as well as an interactive effect that might enhance native-exotic differences (Cruz-Martínez et al. 2009, 2012; Hawkes et al. 2011). Grasslands dominated by exotic species often have lower plant diversity, altered C3-C4 biomass proportions, and different phenologies (Christian and Wilson 1999; Martin et al. 2014; Wilsey and Polley 2006; Wilsey et al. 2009, 2011, 2017; Xu et al. 2015). These changes in the aboveground plant community could have important impacts on belowground nutrient cycling and soil microbial communities as novel ecosystems become more prevalent (Bardgett and van der Putten 2014). Gibbons et al. (2017) studied how microbial communities were impacted by one of three different annual exotic species: Centaurea stoebe (knapweed), B. tectorum, and Euphorbia esula (leafy spurge), and found that exotic species affected soil properties and the structure and composition of microbial communities.
Exotic-dominated plant communities could drive long-term differences in microbial communities by altering the plant carbon (C) to nitrogen (N) ratio in root and shoot litter or the root distribution with depth (Coleman et al. 2000; Wilsey and Polley 2006). This effect may be positive or negative depending on the mode of photosynthesis of the invading plant species’ (Martin et al. 2014). In systems dominated by C3 plant species, mineralization rates are usually increased by exotic species (Stark and Norton 2015; Wedin and Tilman 1992). In systems dominated by C4 grasses or annual species, the invasion of exotic species led to increased litter biomass, C:N, and lignin:N ratios, which reduced mineralization rates (Evans et al. 2001). In our system, the southern plains of the USA, exotic communities are strongly dominated by perennial C4 grasses with high C:N ratios, whereas native communities are diverse mixtures of perennial C3 forbs, C3 grasses, and C4 grasses (Wilsey et al. 2011; Xu et al. 2015). Wilsey and Polley (2006) found that root:shoot ratios, deep root mass, and root mass fraction were lower in exotic than native perennial grasses.
Changes in microorganisms can lead to feedback that affect plant communities by altering nutrient cycling (Berg and Smalla 2009). The plant-microbe-soil feedback loop is theorized to influence a whole range of ecosystem services and dynamics, ranging from litter decomposition to biodiversity-functioning relationships, and natural selection and fitness (Bardgett and van der Putten 2014; Kardol et al. 2013; van der Putten et al. 2013). For example, microbes decompose organic matter and convert N and phosphorous (P) into inorganic forms (mineralization), making them available for plant uptake. Due to the strong relationships between native plants and microorganisms that inhabit all parts of plants (rhizosphere, endosphere, and phyllosphere), as well as those residing in bulk soil, any changes in grassland plant community composition may have strong effects on microbial communities, and in turn, on ecosystem functioning (Bakker et al. 2013; Vandenkoornhuyse et al. 2015). Yet predictive capabilities for how the conversion from native to novel ecosystems will impact microbial community composition, diversity, neutral vs. niche processing, and biogeochemical processes remain unresolved. Microbial changes could be especially pronounced following exotic invasions, because exotic plants are recent arrivals with no evolutionary history with microbes in the invaded range, have differing phenologies than native plants, and could alter nitrogen-fixing microbes due to the variance in tissue types in exotic plant communities.
Replacement of native plant communities by exotic species could alter the structure as well as the composition of microbial communities. Both niche and neutral processes can shape microbial communities (Dini-Andreote et al. 2015; Dumbrell et al. 2010; Morrison-Whittle and Goddard 2015). Niche processes lead to structured microbial communities that reflect compositional variation across host plants, time periods, or soil properties. Neutral processes are random effects (ecological drift) unrelated to habitat conditions that can also lead to variation in microbial communities (Dumbrell et al. 2010; Stegen et al. 2012). If neutral processes are solely operating on microbial communities, then variation should be unrelated to native or exotic plant species shifts or changes in rainfall patterns. However, the influence of native vs. exotic plants or altered rainfall on microbial community structure has not been examined. Exotic plant species are recent arrivals in North America, and they have a shorter history of plant-microbe interactions in this environment compared to native plant species. Based on this, we predicted that neutral processes would dominate in exotic-dominated grasslands, and niche processes would be stronger in native plant communities.
We compared microbial community composition, diversity, and functioning (N and P mineralization) in exotic and native dominated grassland plots, with and without summer irrigation using the long-term Maintenance of Exotic vs. Native Diversity (MEND) experiment in Central Texas (Wilsey et al. 2011). Over time, exotic plant communities developed much lower plant species diversity, higher net primary productivity (NPP), and stronger dominance by C4 grass species than native communities (Wilsey et al. 2011, 2014; Martin et al. 2014). This has led to changes in plant inputs into the soil that could affect microbial composition and functioning. We hypothesized that: 1) bacterial and fungal diversity and community composition differ between exotic and native plant communities, and that this difference is sustained over time, 2) the shifts under exotic vs. native plants will lead to changes in ecosystem functioning (N and P mineralization), 3) native-exotic differences will vary across rainfall treatments, with exotic plot microbial communities responding with greater variability to rainfall inputs, 4) structure of microbial communities and co-occurrence of microbial phylotypes is more impacted by niche than neutral processes in native than exotic communities.
Methods
Study experimental design
The Maintenance of Exotic vs. Native Diversity (MEND) experiment was established early in 2008 at the Grassland, Soil, and Water Research Laboratory (Temple, TX) (Polley et al. 2016; Wilsey et al. 2011, 2014). A detailed description of the experimental design can be found in Wilsey et al. (2011, 2014). Briefly, the experiment was designed as a factorial with 1 m × 1 m plots of mixtures of all native or all exotic plant species crossed with a summer irrigation treatment (128 mm per year added from July 15 to August 15 or no additions as a control) with 16 plots per treatment. The 64 experimental mixtures were planted to identical plant densities, functional group proportions, and species diversity to determine whether differences develop over time between native and exotic plant communities with and without altered precipitation. Either all native species or all exotic species were assigned to 9-species mixtures using a paired species design and species pool of 36 native and exotic species (Table S1). All plant species used were perennials. Replication was achieved with 8 random species draws per treatment and 2 true replicates per random draw and treatment (n = 16 mixtures in each of the four treatments). The 2-m alleyways between plots were seeded with Bouteloua curtipendula (Daneshgar et al. 2013). Plots were not fertilized during the duration of the study.
By October 2009, the plots had stabilized into a high diversity native system and a low diversity exotic-dominated system, which matches closely with the differences between native and exotic-dominated fields in the area (Wilsey et al. 2011, 2014). Exotic plots were strongly dominated by C4 grasses, whereas, native plots had a diverse mixture of C3 forbs and grasses and C4 grass species (Daneshgar et al. 2013; Martin et al. 2014). There were also differences between native and exotic species in aboveground biomass production (peak biomass) and response to summer irrigation (Polley et al. 2014, 2016). Plant communities were sampled in June and October of each year as detailed by Wilsey et al. (2011) and Xu et al. (2015).
N and P mineralization
Polyvinyl chloride (PVC) rings, 10 cm width × 15 cm depth, were pushed into the soil in the center of each plot in March 2015. Rings were kept root free by weeding to prevent root uptake of nutrients (Raison et al. 1987). Thus, the PVC ring provided a barrier to root uptake to separate microbial processes inside the ring from plant+microbial processes outside the ring (Raison et al. 1987). Within each ring, one anion and one cation membrane (Western Ag Innovations, Saskatoon, Saskatchewan, Canada) were inserted on March 16th and removed June 1st, 2015 (Time period 1), inserted again on June 22nd and removed August 18th, 2015 (Time period 2), and inserted on August 25th and removed November 12th, 2015 (Time period 3). These dates were selected to cover the long growing seasons at our field site. Total N mineralization, taken as NO3− and NH4+ accumulation during each time period, was measured colorimetrically using an automated flow injection system. In the third time period, rodents damaged a few cation probes, and total N was calculated with NO3− only. Analyzing the data without these plots did not change the results because NH4+ only contributed a trace amount to total N mineralization. Phosphorus mineralization, PO4−, was measured with an inductively coupled plasma spectrometer (ICP) for each time period.
Soil sample collection for microbial analysis
Two soil cores (3.2 cm diameter) were subsampled from each plot to 10 cm depth in October 2009, 2014, and 2015 at the end of the second, seventh, and eighth growing seasons. The soil cores were stored on dry ice and immediately shipped to Iowa State University (Ames, IA). The soil samples from the two cores per plot per date were composited, homogenized, subsampled, and stored at −80 °C until DNA extraction.
DNA extraction
Microbial DNA was isolated from 0.25 g of soil using the PowerMax Soil DNA Isolation Kit (MoBio Laboratories Inc., Carlsbad, CA) following the manufacturer’s instructions. DNA concentration was determined using a NanoDrop 2000 spectrophotometer (ThermoScientific, Wilmington, DE). The DNA samples were stored at −80 °C until library preparation for sequencing.
Library preparation and amplicon sequencing
Amplicon libraries were prepared and sequenced at Argonne National Laboratory (Lemont, IL). The amplification of V4 region of 16S rRNA for construction of amplicon libraries was performed using primers (515F-806R) and protocol described by Caporaso et al. (2010) for samples collected in 2009, and the modified protocol for samples collected in 2014 and in 2015 (Caporaso et al. 2012). The amplification of Internal Transcribed Spacer (ITS) region for construction of amplicon libraries was performed for samples collected in each year using primers (ITS1f-ITS2) (Smith and Peay 2014) and the sequencing protocol of Caporaso et al. (2012). Following the multiplexing protocol, agarose gel electrophoresis was used to determine the presence of PCR products, and DNA concentration was measured with Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA). The samples were sequenced using sequencing primers for pair-end sequencing. Samples from 2009 (V4 of 16S rRNA) were sequenced on Genome Analyzer IIx (GAIIx, Illumina, San Diego, CA) and samples from 2014 and 2015 were sequenced on MiSeq (Illumina, San Diego, CA).
Sequence processing
Bacterial and fungal sequences were processed through the open access ‘hundo’ v1.1.6 pipeline (https://zenodo.org/record/1193819). Bacterial sequence reads were demultiplexed with EA-Utils (Aronesty 2013) with zero mismatches allowed in the barcode sequence followed by quality filtering with BBDuk2 (Bushnell 2014) for removal of adapter sequences and PhiX with matching kmer length of 31 bp at a hammering distance of 1. Any reads shorter than 51 bp were removed. USEARCH was used to merge reads (Edgar 2010) with a minimum length threshold of 125 bp and maximum error rate of 1%. After sequences were dereplicated (minimum sequence abundance of 2), they were clustered using USEARCH (Edgar 2013) at 97% pairwise sequence identity among operational taxonomic unit (OTU) member sequences. Concurrently, de novo prediction of chimeric sequences was performed during clustering. BLAST was used to assigned taxonomy to OTU sequence (Camacho et al. 2009) alignments followed by least common ancestor assignments across SILVA database version 123 clustered at 99% (Quast et al. 2013). OTU seed sequences were filtered against SILVA database version 123 clustered at 99% to identify chimeric OTUs using USEARCH.
Fungal sequences were processed using the same approach with a few differences: 175 bp as a minimum length threshold and UNITE v. 7 database for assigning the taxonomy and identification of chimeric OTUs. In both cases, sequences were aligned using Clustal Omega (Sievers et al. 2011) and FastTree2.0 (Price et al. 2010) was used to generate the phylogenetic tree in a Newick format.
For 16S sequences, any non-prokaryotic or phylum-unassigned sequences were removed from the OTU table and a total number of sequences for all three years for bacteria was 11,839,740 (2009: 8,033,517; 2014: 1,949,334; 2015: 1,856,889). For ITS sequences, unassigned sequences were removed and the total number of sequences for all three years for fungi was 4,317,020 (2009: 1,344,230; 2014: 826,862; 2015: 2,145,928). The sequences were assigned across 20,804 bacterial and 3280 fungal OTUs.
Statistical analyses
The preliminary statistical analysis was conducted in QIIME using “core diversity” script (Caporaso et al. 2010). The number of sequences per sample was normalized by rarefaction to the lowest number of reads in bacterial and fungal libraries (4306 for bacteria and 5613 for fungi). Subsampling a given population allows for the comparison of multiple samples, that otherwise varies in abundance because of the sequencing process. The normalized (rarefied) data were used in all statistical tests.
Microbial diversity analysis
Alpha diversity was estimated by calculating Simpson’s reciprocal (1/∑pi2), Faith’s phylogenetic diversity (PD) indexes, and Chao1 richness estimator. To determine the impact of native and exotic plant treatments and irrigation on microbial diversity measures, a mixed model ANOVA (Proc MIXED), with main effects of plant treatments (Native vs. Exotic) and irrigation (128 mm or 0 mm) as fixed effects, and draw and rep as random terms was performed. Repeated measures analyses (diversity measures) used first-order autoregressive (AR(1)) covariance structure calculations in SAS 9.4 (SAS Institute, Cary, NC).
Microbial community composition analysis
To determine how plant treatments, irrigation, and their interaction shaped microbial community composition, R Studio v. 1.0.136 was used to perform permutation-based non-parametric multivariate analysis of variance on microbial OTU abundance using ‘Vegan’ package. OTU abundance was used for a PERMANOVA analysis, using a two-way factorial design (plant treatment, summer irrigation, and plant and irrigation treatments) with 999 permutations for each year. Samples that had low read counts, <1000 reads, were removed from the analysis and the corresponding draws were removed from PERMANOVA. Additionally, a non-metric multidimensional scaling (NMDS) analysis using a Bray-Curtis dissimilarity matrices was performed with plant treatments (native vs. exotic), summer irrigation, and the interaction between plant treatments and irrigation, to determine their impact on OTU abundance, microbial composition. Following, NMDS scree plots were used to determine the number of dimensions for presenting each ordination. The normalized data were filtered to contain only OTUs representing at least 0.1% of total reads in each sample using QIIME (“filter otus from otu table” command).
Niche and neutral processes analyses
We determined the influence of both neutral and niche processes on microbial community structure under native vs. exotic plants by using checkerboard scores (C-scores). The C-score (Stone and Roberts 1990) was calculated in EcoSim v. 7.0 (Gotelli and Entsminger 2004) as a presence/absence matrix on normalized OTU table that was previously filtered (only including OTUs with >0.1% absolute abundance). The C-score determines the strength of this non-random co-occurrence: the higher the C-score, more segregation and presumably, competition exist between species (niche differentiation). If the C-score is low, species co-exist together without competition (neutral processes dominate). Another measure of significant non-random co-occurrence is the standardized effect size (SES). SES is significant at values below −2 or above 2. Statistical significance was based on a permutation procedure comparing calculated C scores to randomly derived C scores.
Microbial functional group analyses
To evaluate unique OTUs to each plant treatment, the group significance command with Kruskal-Wallis test to estimate mean abundance of individual OTUs in native and exotic plots, with the Benjamini-Hochberg false discovery rate (FDR) correction applied to p values. Functional trophic modes were analyzed for the differentially abundant fungal OTUs within native and exotic samples by using FUNguild (https://github.com/UMNFuN/FUNGuild) (Nguyen et al. 2016).
N and P mineralization analyses
A mixed model ANOVA was used to determine the impact of plant treatment (exotic vs. native) and summer irrigation on microbial mineralization of N and P. A mixed model ANOVA was performed using plant treatment and irrigation as fixed effects and draw and rep as random terms. Mineralization data was analyzed as a repeated measure and first-order autoregressive covariance structure was used in calculations. To determine if particular microbial community members had a significant correlation with N or P mineralization, an NMDS biplot with cutoff R2 value = 0.2, was performed on microbial community data from 2015 (the same year as mineralization) and average total N and P mineralization rates.
Results
Microbial community composition
The majority of bacterial sequences were assigned at the phylum level at a minimum (99.75%). Dominant bacterial phyla included: Proteobacteria (35%), Acidobacteria (19.3%), Bacteroidetes (17.3%), Actinobacteria (9.5%), and Verrucomicrobia (8.9%) (Table S2). Fungal communities were dominated by the Ascomycota (65.5%) and Zygomycota (14.8%) phyla, with less abundant phyla being unclassified phyla (9.4%), Basidiomycota (9%) and Glomeromycota (0.11%).
Plant treatment but not irrigation had a strong effect on fungal community composition at the OTU level across the years (factorial PERMANOVA, P < 0.05) (Table 1). There was no significant interaction effect of plant treatment and irrigation except in 2015 (P = 0.015). As demonstrated in the NMDS plots, differences in community structure were driven primarily by plant treatment (P < 0.05), however, the differences between native and exotic communities were relatively small (Fig. 1).
Fungal non-metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarity distance measures for OTU abundance by plant treatment (native vs. exotic) by sampling year, axis 1 vs. 2 and axis 1 vs. 3 are shown, a) 2009 (2D stress 0.229, 3D stress 0.171), b) 2014 (2D stress 0.253, 3D stress 0.182), c) 2015 (2D stress 0.243, 3D stress 0.182)
To further evaluate the impact of plant treatment on the fungal community composition, we performed a differential abundance test between the native and exotic plant community plots. The testing was not performed for summer irrigation treatment because of no effect on microbial communities as determined by PERMANOVA. Fourteen fungal OTUs differed in their abundance between native and exotic plots (P < 0.05 after Benjamini-Hochberg FDR correction) (Table 2) and were classified based on the trophic mode: saprotrophs, pathogens, or the OTUs with unknown function. Among the listed OTUs, only four were identified to the genus level: Stagonospora, Humicola grisea var. grisea, Lophiostoma and Basidioascus undulatus. OTU representing Stagonospora genus was assigned to the pathogen group, and it was more abundant in native plots. Saprotrophs were represented by six OTUs. One OTU of order Hypocreales was more abundant in exotic plots. The remaining OTUs are unknown in their trophic mode by FUNguild analysis. Unlike the fungal community, no bacterial OTUs were found to be significantly different in their mean abundance between native and exotic plots (Kruskal-Wallis test, P > 0.05 after Benjamini-Hochberg FDR correction).
Two-way factorial PERMANOVA showed an effect of plant treatment on bacterial community composition in 2009 and 2015 (P < 0.05). The summer irrigation treatment had a marginal effect on the bacterial community only in 2009, which was the second growing season. No interactions effects of plant treatment with summer irrigation were detected for the bacterial community. NMDS revealed patterns consistent with PERMANOVA results showing significant sample clustering based on plant treatment in 2009 and 2015 although similarly to fungi the differences were relatively small (Fig. 2).
Bacterial non-metric multidimensional scaling (NMDS) based on Bray-Curtis dissimilarity distance measures for OTU abundance by plant treatment (native vs. exotic) by sampling year, axis 1 vs. 2 and axis 1 vs. 3 are shown, a) 2009 (2D stress 0.187, 3D stress 0.130), b) 2014 (2D stress 0.149, 3D stress 0.113), c) 2015 (2D stress 0.226, 3D stress 0.160)
Bacterial OTU differential abundance was also analyzed to determine the impact of the plant treatment on the bacterial community. However, unlike the fungal community, no bacterial OTUs were found to be significantly different in their mean abundance between native and exotic plots (Kruskal-Wallis test, P > 0.05 after Benjamini-Hochberg FDR correction).
Microbial diversity
Diversity measurements for the microbial community were analyzed to determine if plant treatment or irrigation had an impact on the diversity of the microbial community. Fungal diversity measures, Chao1 and Faith’s phylogenetic diversity, were similar between plant treatment and summer irrigation treatments, and the interaction of the plant treatment and summer irrigation was not significant (Fig. S1). However, there was a large drop in fungal diversity in 2014 compared to sampling years, 2009 and 2015. The interaction between plant treatment and summer irrigation moderately affected fungal Simpson’s reciprocal, but plant treatment and summer irrigation did not (interaction: F1,185 = 3.542, P = 0.061).
On the other hand, bacterial diversity measures did differ between native and exotic plots, with Chao1 richness and Faith’s phylogenetic diversity showing significantly higher values for exotic than native mixtures (F1,185 = 10.18, P = 0.015 and F1,185 = 7.85, P = 0.027, respectively). Faith’s phylogenetic diversity was much higher during 2009 than 2014 and 2015. However, Chao1 richness had the opposite pattern, with lower richness values in 2009 than in 2014 and 2015. Simpson’s diversity measures (Fig. S2) were not statistically significant for any treatment.
Microbial functioning: mineralization rates in native vs. exotic mixtures
Microbial functioning was assessed as mineralization of N and P. Nitrogen mineralization was significantly higher under native than exotic plots (F1,27 = 34.2, P < 0.001). This was consistent across time periods and irrigation treatments (i.e., there was no time x plant treatment interaction). Nitrogen mineralization was lowest during the middle (warmest) part of the growing season (Time F2,118 = 41.2, P < 0.001) (Fig. 3a).
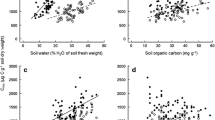
Phosphorus mineralization was greater in native than exotic plots only at the final sampling (plant treatment F1,59 = 15.09, P = 0.0003; time x plant treatment interaction F2,118 = 9.02, P = 0.0002). Phosphorus mineralization was significantly lower in exotic plots in the last sampling period (Fig. 3b). Several fungal taxa were associated with the change in mineralization (Fig. 4). Increased N and P mineralization were associated with fungal OTUs representing Humicola grisea var. grisea, order Mortierellales.
Niche vs. neutral processed in microbial communities
The checkerboard score (C-score) was calculated for the co-occurrence analysis in native vs. exotic plots to determine if niche or neutral processes predominated. If the C-score for the observed data is significantly different than the C-score for simulated data, species occur non-randomly (e.g., communities are more structured). Generally, in both native and exotic plots, non-random co-occurrence was observed (Pobs < =Psim) (Table 3). The C-scores were much higher for fungal than bacterial communities, implying less randomness, and more niche differentiation among fungi than bacteria. C-scores were higher in exotic than native plots for both bacterial and fungal communities, suggesting greater structure in microbial communities under exotic than native plant communities.
Discussion
We conducted an eight-year study of microbial communities in soils beneath native and exotic plant species mixtures that either were exposed to summer irrigation or were non-irrigated controls. In this highly replicated experiment that was established in 2008 and sampled through 2015, we observed that composition, structure, and function of microbial communities differed between native and exotic plant communities. Summer irrigation did not significantly change the microbial community structure. Fungal community composition was more strongly affected by native vs. exotic plant plots than was bacterial community composition. The differences in composition were driven through the differential abundance of certain fungal phylotypes, among which a pathogen and saprotrophs were detected, as well as some with unknown function. This coincided with differences in function, with lower N and P mineralization in exotic than native plots. Finally, fungal community composition was more non-random in co-occurrence in plots of exotic than native plant communities.
We hypothesized that because native plant-microbe communities co-evolved together they would be more structured than microbial communities under exotic plants. Notably, we found support for the opposite outcome; exotic plots had stronger niche processes than native plots. As the MEND experiment progressed over time, native and exotic plots diverged from each other in terms of diversity, with lower plant diversity in exotic plots. The exotic plots were strongly dominated by various C4 grasses, whereas native plots contained mixtures of C3 and C4 grasses and C3 forbs (Wilsey et al. 2011, 2014; Martin et al. 2014). The microbial community composition and diversity differed between exotic and native plots as well. These plant differences could have had a stronger filtering effect on the microbial community in exotic plots generating higher incidences of non-random co-occurrence. Fungi, in particular, had higher non-random co-occurrence than bacterial communities in exotic than native plots.
Our results show bacterial communities were more impacted by neutral processes compared to fungal communities, which were more strongly impacted by niche processes. Due to the differences in spatial scale of a bacterium’s habitat vs. that of a fungus, it is often more common to find differences in fungal community processes when studied at relatively large spatial scales, e.g. bulk soil compared to targeted studies, e.g. rhizosphere or soil aggregates (Bach et al. 2018; Nemergut et al. 2013). Further exploration of this relationship between rhizosphere and bulk soil in native and exotic plots can test whether the occurrence is even less random due to varying fungal trophic modes. In addition, use of this finer spatial scale design may reveal stronger bacterial roles within changes between native and exotic plant community interactions, to account for their smaller physical size.
Plant treatment (native vs. exotic plots), also impacted fungal community phylotype abundance. The plant pathogen genus Stagonospora (Solomon et al. 2006) was significantly more abundant in native plots. This higher accumulation of the genetic signatures of this genus in native plots compared with exotic plots might suggest a ready accumulation of pathogens in native plants. Similarly, we detected a few fungal OTUs belonging to saprotrophic groups and the majority had unclassified functions. The majority of environmental microorganisms are unculturable, consequently most sequences identified organisms have unknown ecological roles; as our understanding of individual fungi’s physiology increases so will our determination of their ecology relevance (Rondon et al. 1999; Vartoukian et al. 2010).
Native and exotic plant communities had distinctly different microbial communities with altered microbial mineralization rates. Microorganisms have been shown to directly and indirectly influence plant communities via mineralization (Schimel and Bennett 2004). Mineralization rates were much higher in native than exotic plots. Native mixtures were more diverse, with a mixture of C3 and C4 plant species present, which would lead to higher substrate quality entering the soil compared to the C4 grass dominated exotic plots (Martin et al. 2014; Xu et al. 2015). C4 grasses have lower N content than C3 species, and this potentially was responsible for the reduced N mineralization in exotic plots. An increase in microbial mineralization has been linked to greater plant biomass as well as increases in tolerance of drought (Panke-Buisse et al. 2015; Treseder and Lennon 2015). The lower mineralization rates under exotic plots may have been constraining their growth response over time, reducing the differences in productivity between native and exotic plots. Under native plant communities, microorganisms provided greater ecosystem services in the plant-microbe-soil feedback loop.
Fungal community changes due to plant treatment were associated with changes in mineralization of N and P. In particular, the abundance of fungal class Agaricostilbomycetes, known saprotrophs, was positively correlated with mineralization rates. Agaricostilbomycetes may represent a key group in this system that regulates mineralization rates of N and P, and this should be tested in the future with more direct manipulations. N and P mineralization rates, in turn, could be affecting plant productivity and both plant and microbial community composition and diversity.
Unlike native-exotic plant differences, we found surprisingly small and non-significant effects of summer irrigation. Few have studied the direct influence of altered precipitation on fungal and bacterial communities. Cruz-Martínez et al. (2009, 2012) found that microbial communities changed within days after rainfall manipulations, showing a pulse response, but remained unchanged over longer multi-year time periods. The short-term change was associated with the relative abundance of the most abundant phyla. Hawkes et al. (2011) investigated how fungal communities responded to periods of altered precipitation and also found the impact to be time-related, with less diverse fungal communities during wetter periods immediately following a rainfall event, but long-term effects were smaller. However, the long-term effects of rainfall events vary, dependent upon the magnitude of change in the aboveground community (Cruz-Martínez et al. 2009). In our study, we did not see any direct effects of irrigation, however, we do see potentially indirect impacts due to changes in plant communities within both exotic and native plots. Irrigation may have a direct short-term impact on the microbial community function, however, the long-term impacts can be mediated by the aboveground community.
We did have large effects on microbial communities from year to year differences, especially in sampling years 2014 vs. 2015. Both years were wet, but 2014 was cooler than average (Xu et al. 2017). Temperature may have had important effects on microbial communities, and the effects of temperature on microbial in grasslands deserve more study.
In summary, fungal community composition and C-scores differed between diverse native and exotic C4 grass dominated plots. Bacterial communities were not as strongly affected by plant community treatments. The fungal communities were more structured in exotic than native plots, potentially due to the simplification of the aboveground plant community. We also showed that nutrient cycling for N and P was higher under the more diverse C3-C4 mixed native than the low diversity, C4 dominated exotic mixtures. The changes in plant production and functional group abundance that characterized exotic compared to native plant communities had a significant impact on the belowground microbial community structure, composition, and function. Our study demonstrates that exotic plants can have cascading impacts on ecosystems by changing microbial community composition and biogeochemical cycles.
References
Allan RP, Soden BJ (2008) Atmospheric warming and the amplification of precipitation extremes. Science 80:1481–1484
Aronesty E (2013) Comparison of sequencing utility programs. Open Bioinforma J 7:1–8
Bach EM, Williams RJ, Hargreaves SK, Yang F, Hofmockel KS (2018) Greatest soil microbial diversity found in micro-habitats. Soil Biol Biochem 118:217–226
Bakker PA, Berendsen RL, Doornbos RF, Wintermans PC, Pieterse CM (2013) The rhizosphere revisited: root microbiomics. Front Plant Sci 4:165
Bardgett RD, van der Putten WH (2014) Belowground biodiversity and ecosystem functioning. Nature 515:505–511
Berg G, Smalla K (2009) Plant species and soil type cooperatively shape the structure and function of microbial communities in the rhizosphere. FEMS Microbiol Ecol 68:1–13
Bushnell B (2014) BBMap: A fast, accurate. In: Splice-aware aligner
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K et al (2009) BLAST+: architecture and applications. BMC Bioinformatics 10:1–9
Caporaso JG, Lauber CL, Walters WA, Berg-lyons D, Lozupone CA, Turnbaugh PJ et al (2010) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A 108:4516–4522
Caporaso JG, Lauber CL, Walters WA, Berg-lyons D, Huntley J, Fierer N et al (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J 6:1621–1624
Christian JM, Wilson SD (1999) Long-term ecosystem impacts of an introduced grass in the northern Great Plains. Ecology 80:2397–2407
Coleman MD, Dickson RE, Isebrands JG (2000) Contrasting fine-root production , survival and soil CO2 efflux in pine and poplar plantations. Plant Soil 225:129–139
Costa, I, Costa Assuncao, M, Lima, TEF de, Oliveira, RJV, Queiroz (2012) Checklist of endophytic fungi from tropical regions. Mycotaxon 119:1–26
Cruz-Martínez K, Suttle KB, Brodie EL, Power ME, Andersen GL, Banfield JF (2009) Despite strong seasonal responses, soil microbial consortia are more resilient to long-term changes in rainfall than overlying grassland. ISME J. 3:738–744
Cruz-Martínez K, Rosling A, Zhang Y, Song M, Andersen GL, Banfield JF (2012) Effect of rainfall-induced soil geochemistry dynamics on grassland. Appl Environ Microbiol 78:7587–7595
Cunfer BM (2000) Stagonospora and Septoria diseases of barley, oat, and rye. Can J Plant Pathol 22:332–348
Daneshgar PP, Polley HW, Wilsey BJ (2013) Simple plant traits explain functional group diversity decline in novel grassland communities of Texas. Plant Ecol 214:231–241
van der Putten WH, Bardgett RD, Bever JD, Bezemer TM, Casper BB, Fukami T, Kardol P, Klironomos JN, Kulmatiski A, Schweitzer JA, Suding KN (2013) Plant–soil feedbacks: the past, the present and future challenges. J Ecol 101:265–276
Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF (2015) Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci 112:E1326–E1332
Dukes JS, Mooney HA (1999) Does global change increase the success of biological invaders ? TREE 14:135–139
Dumbrell AJ, Nelson M, Helgason T, Dytham C, Fitter AH (2010) Relative roles of niche and neutral processes in structuring a soil microbial community. The ISME journal 4:337–345
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10:996–998
Evans RD, Rimer R, Sperry LJ, Belnap J (2001) Exotic plant invasion alters nitrogen dynamics in an arid grassland. Ecol Appl 11:1301–1310
Gibbons SM, Lekberg Y, Mummey DL, Sangwan N, Ramsey PW, Gilbert JA (2017) Invasive plants rapidly reshape soil properties in a grassland ecosystem. mSystems 2:e00178–e00116
Gotelli, NJ, Entsminger, GL (2004) EcoSim: Null models software for ecology
Hawkes CV, Kivlin SN, Rocca JD, Huguet V, Thomsen MA, Suttle KB (2011) Fungal community responses to precipitation. Glob Chang Biol 17:1637–1645
Hobbs RJ, Arico S, Aronson J, Baron JS, Bridgewater P, Cramer VA, Epstein PR, Ewel JJ, Klink CA, Lugo AE, Norton D, Ojima D, Richardson DM, Sanderson EW, Valladares F, Vila M, Zamora R, Zobel M (2006) Novel ecosystems: theoretical and management aspects of the new ecological world order. Glob Ecol Biogeogr 15:1–7
Hobbs RJ, Higgs E, Harris JA (2009) Novel ecosystems: implications for conservation and restoration. Trends Ecol Evol 24:599–605
Hobbs RJ, Higgs ES, Hall CM (2013) Introduction: why novel ecosystems? In: Novel ecosystems: intervening in the new ecological world order, pp 1:1–1:8
Kardol P, Deyn GB, Laliberte E, Mariotte P, Hawkes CV (2013) Biotic plant–soil feedbacks across temporal scales. J Ecol 101:309–315
Martin LM, Polley HW, Daneshgar PP, Harris MA, Wilsey BJ (2014) Biodiversity, photosynthetic mode, and ecosystem services differ between native and novel ecosystems. Oecologia 175:687–697
Morrison-Whittle P, Goddard MR (2015) Quantifying the relative roles of selective and neutral processes in defining eukaryotic microbial communities. The ISME journal 2015
Nemergut DR, Schmidt SK, Fukami T, O'Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S (2013) Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356
Nguyen NH, Song Z, Bates ST, Branco SS, Tedersoo L, Menke J et al (2016) FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol 20:241–248
Panke-Buisse K, Poole AC, Goodrich JK, Ley RE, Kao-Kniffin J (2015) Selection on soil microbiomes reveals reproducible impacts on plant function. The ISME journal 9:980–989
Polley HW, Derner JD, Jackson RB, Wilsey BJ, Fay PA (2014) Impacts of climate change drivers on C4 grassland productivity: scaling driver effects through the plant community. J Exp Bot 65:3415–3424
Polley HW, Gibson AE, Fay PA, Wilsey BJ (2016) Biotic regulation of CO2 uptake – climate responses: links to vegetation properties. Ecosystems 19:1376–1385
Price MN, Dehal PS, Arkin AP (2010) FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:1–10
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:590–596
Raison RJ, Connel MJ, Khanna PK (1987) Methodology for studying fluxes of soil mineral-N in situ. Soil Biol Biochem 19:521–530
Rondon MR, Goodman RM, Handelsman J (1999) The Earth’s bounty: assessing and accessing soil microbial diversity. Trends Biotechnol 17:403–409
Schimel JP, Bennett J (2004) Nitrogen mineralization: challenges of a changing paradigm. Ecology 85:591–602
Seastedt TR, Hobbs RJ, Suding KN (2008) Management of novel ecosystems: are novel approaches required? Front Ecol Environ 6:547–553
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W et al (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal omega. Mol Syst Biol 7:1–6
Smith DP, Peay KG (2014) Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS One 9:1–12
Solomon PS, Lowe RGT, Tan KC, Waters ODC, Oliver RP (2006) Stagonospora nodorum: cause of stagonospora nodorum blotch of wheat. Mol Plant Pathol 7:147–156
Stark JM, Norton JM (2015) The invasive annual cheatgrass increases nitrogen availability in 24-year-old replicated field plots. Oecologia 177:799–809
Stegen JC, Lin X, Konopka AE, Fredrickson JK (2012) Stochastic and deterministic assembly processes in subsurface microbial communities. The ISME journal 6:1653–1664
Sterkenburg E, Bahr A, Brandström Durling M, Clemmensen KE, Lindahl BD (2015) Changes in fungal communities along a boreal forest soil fertility gradient. New Phytol 207:1145–1158
Stone L, Roberts A (1990) The checkerboard score and species distributions. Oecologia 85:74–79
Tedersoo L, Bahram M, Põlme S, Kõljalg U, Yorou NS, Wijesundera R et al (2014) Global diversity and geography of soil fungi. Science 80:346
Treseder KK, Lennon JT (2015) Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev 79:243–262
Vandenkoornhuyse P, Quaiser A, Duhamel M, Le Van A, Dufresne A (2015) The importance of the microbiome of the plant holobiont. New Phytol 206:1196–1206
Vartoukian SR, Palmer RM, Wade WG (2010) Strategies for culture of ‘unculturable’bacteria. FEMS Microbiol Lett 309:1–7
Wedin, DA, Tilman, D (1992). Nitrogen cycling, plant competition, and the stability of tallgrass prairie. In Proceedings of the Twelfth North American Prairie Conference. University of Northern Iowa Press, Cedar Falls, Iowa 5–8
Wilsey BJ, Polley HW (2006) Aboveground productivity and root-shoot allocation differ between native and introduced grass species. Oecologica 150:300–309
Wilsey BJ, Teaschner TB, Daneshgar PP, Isbell FI, Polley HW (2009) Biodiversity maintenance mechanisms differ between native and novel exotic-dominated communities. Ecol. Lett 12:432–442
Wilsey BJ, Daneshgar PP, Polley HW (2011) Biodiversity, phenology and temporal niche differences between native- and novel exotic-dominated grasslands. Perspect Plant Ecol Evol Syst 13:265–276
Wilsey BJ, Daneshgar PP, Hofmockel KS, Polley HW (2014) Invaded grassland communities have altered stability-maintenance mechanisms but equal stability compared to native communities. Ecol Lett 17:92–100
Wilsey BJ, Martin LM, Kaul AD (2017) Phenology differences between native and novel exotic-dominated grasslands rival the effects of climate change. J Appl Ecol in press. https://doi.org/10.1111/1365-2664-1291
Xu X, Polley HW, Hofmockel KS, Daneshgar PP, Wilsey BJ (2015) Plant invasions differentially affected by diversity and dominant species in native- and exotic-dominated grasslands. Ecol Evol 5:5662–5670
Xu, X, Polley, HW, Hofmockel, K, Wilsey, BJ, (2017) Species composition but not diversity explains recovery from the 2011 drought in Texas grasslands. Ecosphere, 8:1:11
Acknowledgments
We thank USDA – NIFA grant 2014-67003-22067 and NSF DEB-0639417 for funding support. We also thank Emily Powers, Katherine Jones, and Chris Kolodziejczyk for their work in maintaining the MEND research site. In addition, we would like to thank numerous undergraduate research assistants for their support both in Texas and Iowa.
Data deposition
The raw sequences as FASTQ files were deposited in NCBI Sequence Read Archive (SRA) under no. SRP115489.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Responsible Editor: Paul Bodelier.
Electronic supplementary material
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Checinska Sielaff, A., Upton, R.N., Hofmockel, K. et al. Microbial community structure and functions differ between native and novel (exotic-dominated) grassland ecosystems in an 8-year experiment. Plant Soil 432, 359–372 (2018). https://doi.org/10.1007/s11104-018-3796-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11104-018-3796-1