Abstract
Key message
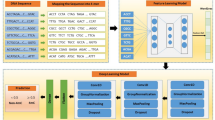
This study used k-mer embeddings as effective feature to identify DNA N6-Methyladenine sites in plant genomes and obtained improved performance without substantial effort in feature extraction, combination and selection.
Abstract
Identification of DNA N6-methyladenine sites has been a very active topic of computational biology due to the unavailability of suitable methods to identify them accurately, especially in plants. Substantial results were obtained with a great effort put in extracting, heuristic searching, or fusing a diverse types of features, not to mention a feature selection step. In this study, we regarded DNA sequences as textual information and employed natural language processing techniques to decipher hidden biological meanings from those sequences. In other words, we considered DNA, the human life book, as a book corpus for training DNA language models. K-mer embeddings then were generated from these language models to be used in machine learning prediction models. Skip-gram neural networks were the base of the language models and ensemble tree-based algorithms were the machine learning algorithms for prediction models. We trained the prediction model on Rosaceae genome dataset and performed a comprehensive test on 3 plant genome datasets. Our proposed method shows promising performance with AUC performance approaching an ideal value on Rosaceae dataset (0.99), a high score on Rice dataset (0.95) and improved performance on Rice dataset while enjoying an elegant, yet efficient feature extraction process.


Similar content being viewed by others
Data availability
The data is published and specified in the paper.
Code availability
Source code is provided at https://github.com/khucnam/Deep_Emb_6mA.
References
Basith S, Manavalan B, Shin TH, Lee G (2019) SDM6A: a web-based integrative machine-learning framework for predicting 6mA sites in the Rice genome. Mol Ther Nucleic Acids 18:131–141
Bojanowski P, Grave E, Joulin A, Mikolov T (2017) Enriching word vectors with subword information. Trans Assoc Comput Linguist 5:135–146
Chen W, Lv H, Nie F, Lin H (2019) i6mA-Pred: identifying DNA N6-methyladenine sites in the Rice genome. Bioinformatics 35:2796–2800
Clough E, Barrett T (2016) The gene expression omnibus database. Statistical genomics. Springer, New York, pp 93–110
Fang G, Munera D, Friedman DI, Mandlik A, Chao MC, Banerjee O, Feng Z, Losic B, Mahajan MC, Jabado OJ (2012) Genome-wide mapping of methylated adenine residues in pathogenic Escherichia coli using single-molecule real-time sequencing. Nat Biotechnol 30:1232–1239
Feng P, Yang H, Ding H, Lin H, Chen W, Chou K-C (2019) iDNA6mA-PseKNC: Identifying DNA N6-methyladenosine sites by incorporating nucleotide physicochemical properties into PseKNC. Genomics 111:96–102
Greer EL, Blanco MA, Gu L, Sendinc E, Liu J, Aristizábal-Corrales D, Hsu C-H, Aravind L, He C, Shi Y (2015) DNA methylation on N6-adenine in C. elegans. Cell 161:868–878
Hasan MM, Basith S, Khatun MS, Lee G, Manavalan B, Kurata H (2020a) Meta-i6mA: an interspecies predictor for identifying DNA N6-methyladenine sites of plant genomes by exploiting informative features in an integrative machine-learning framework. Brief Bioinform. https://doi.org/10.1093/bib/bbaa202
Hasan MM, Manavalan B, Shoombuatong W, Khatun MS, Kurata H (2020b) i6mA-Fuse: improved and robust prediction of DNA 6 mA sites in the Rosaceae genome by fusing multiple feature representation. Plant Mol Biol 103:225–234
Joulin A, Grave E, Bojanowski P, Mikolov T (2016) Bag of tricks for efficient text classification. arXiv preprint arXiv:1607.01759
Joulin A, Grave E, Bojanowski P, Douze M, Jégou H, Mikolov T (2016) Fasttext. zip: compressing text classification models. arXiv preprint arXiv:1612.03651
Karanthamalai J, Chodon A, Chauhan S, Pandi G (2020) DNA N6-methyladenine modification in plant genomes—a glimpse into emerging epigenetic code. Plants 9:247
Khanal J, Lim DY, Tayara H, Chong KT (2020) i6mA-stack: a stacking ensemble-based computational prediction of DNA N6-methyladenine (6mA) sites in the Rosaceae genome. Genomics. https://doi.org/10.1016/j.ygeno.2020.09.054
Khanal J, Lim DY, Tayara H, Chong KT (2021) i6ma-stack: a stacking ensemble-based computational prediction of DNA N6-methyladenine (6ma) sites in the Rosaceae genome. Genomics 113:582–592
Liu Z-Y, Xing J-F, Chen W, Luan M-W, Xie R, Huang J, Xie S-Q, Xiao C-L (2019) MDR: an integrative DNA N6-methyladenine and N4-methylcytosine modification database for Rosaceae. Horticult Res 6:1–7
Luo G-Z, Blanco MA, Greer EL, He C, Shi Y (2015) DNA N 6-methyladenine: a new epigenetic mark in eukaryotes? Nat Rev Mol Cell Biol 16:705–710
Luo G-Z, Wang F, Weng X, Chen K, Hao Z, Yu M, Deng X, Liu J, He C (2016) Characterization of eukaryotic DNA N 6-methyladenine by a highly sensitive restriction enzyme-assisted sequencing. Nat Commun 7:1–6
Mikolov T, Sutskever I, Chen K, Corrado GS, Dean J (2013) Distributed representations of words and phrases and their compositionality. In: Advances in neural information processing systems, pp. 3111–3119
O’Brown ZK, Greer EL (2016) N6-methyladenine: a conserved and dynamic DNA mark. DNA methyltransferases-role and function. Springer, Cham, pp 213–246
Pian C, Zhang G, Li F, Fan X (2020) MM-6mAPred: identifying DNA N6-methyladenine sites based on Markov model. Bioinformatics 36:388–392
Pukkila PJ, Peterson J, Herman G, Modrich P, Meselson M (1983) Effects of high levels of DNA adenine methylation on methyl-directed mismatch repair in Escherichia coli. Genetics 104:571–582
Ratel D, Ravanat JL, Berger F, Wion D (2006) N6-methyladenine: the other methylated base of DNA. BioEssays 28:309–315
Roberts D, Hoopes B, McClure W, Kleckner N (1985) IS10 transposition is regulated by DNA adenine methylation. Cell 43:117–130
Smith ZD, Meissner A (2013) DNA methylation: roles in mammalian development. Nat Rev Genet 14:204–220
Tahir M, Tayara H, Chong KT (2019) iDNA6mA (5-step rule): Identification of DNA N6-methyladenine sites in the Rice genome by intelligent computational model via Chou’s 5-step rule. Chemom Intell Lab Syst 189:96–101
Wang X, Yan R (2018) RFAthM6A: a new tool for predicting m 6 A sites in Arabidopsis thaliana. Plant Mol Biol 96:327–337
Xu H, Hu R, Jia P, Zhao Z (2020) 6mA-Finder: a novel online tool for predicting DNA N6-methyladenine sites in genomes. Bioinformatics 36:3257–3259
Ye P, Luan Y, Chen K, Liu Y, Xiao C, Xie Z (2016) MethSMRT: an integrative database for DNA N6-methyladenine and N4-methylcytosine generated by single-molecular real-time sequencing. Nucl Acids Res 45:85–89
Yu N, Li Z, Yu Z (2018) Survey on encoding schemes for genomic data representation and feature learning—from signal processing to machine learning. Big Data Min Anal 1:191–210
Zhang G, Huang H, Liu D, Cheng Y, Liu X, Zhang W, Yin R, Zhang D, Zhang P, Liu J (2015) N6-methyladenine DNA modification in Drosophila. Cell 161:893–906
Zhang M, Sun J-W, Liu Z, Ren M-W, Shen H-B, Yu D-J (2016) Improving N6-methyladenosine site prediction with heuristic selection of nucleotide physical–chemical properties. Anal Biochem 508:104–113
Zhang Q, Liang Z, Cui X, Ji C, Li Y, Zhang P, Liu J, Riaz A, Yao P, Liu M (2018) N6-Methyladenine DNA methylation in Japonica and Indica Rice genomes and its association with gene expression, plant development, and stress responses. Mol Plant 11:1492–1508
Zhou C, Wang C, Liu H, Zhou Q, Liu Q, Guo Y, Peng T, Song J, Zhang J, Chen L (2018) Identification and analysis of adenine N 6-methylation sites in the Rice genome. Nat Plants 4:554–563
Funding
This work was partially supported by the Ministry of Science and Technology, Taiwan, R.O.C. under Grant No. MOST 109-2811-E-155-505 and No. MOST 109-2221-E-155-045.
Author information
Authors and Affiliations
Contributions
Conceptualization, NTTD, TVN, NQKL and YYO; methodology, NTTD and NQKL; formal analysis, TTDN; writing—original draft preparation, TTDN; writing—review and editing, TTDN, TVN, NQKL, and YYO; supervision, YYO; funding acquisition, YYO. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Consent to participate
Not applicable.
Consent for publication
All authors have read and agreed to the published version of the manuscript.
Ethical approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nguyen, T.T.D., Trinh, V.N., Le, N.Q.K. et al. Using k-mer embeddings learned from a Skip-gram based neural network for building a cross-species DNA N6-methyladenine site prediction model. Plant Mol Biol 107, 533–542 (2021). https://doi.org/10.1007/s11103-021-01204-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01204-1