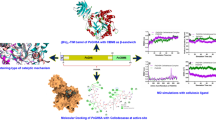
Abstract
Key message
NADP-ME2 from Arabidopsis thaliana exhibits a distinctive and complex regulation by fumarate, acting as an activator or an inhibitor according to substrate and effector concentrations. In this work, we used molecular modeling approach and site-directed mutagenesis to characterized the NADP-ME2 structural determinants necessary for allosteric regulation providing new insights for enzyme optimization.
Abstract
Structure–function studies contribute to deciphering how small modifications in the primary structure could introduce desirable characteristics into enzymes without affecting its overall functioning. Malic enzymes (ME) are ubiquitous and responsible for a wide variety of functions. The availability of a high number of ME crystal structures from different species facilitates comparisons between sequence and structure. Specifically, the structural determinants necessary for fumarate allosteric regulation of ME has been of particular interest. NADP-ME2 from Arabidopsis thaliana exhibits a distinctive and complex regulation by fumarate, acting as an activator or an inhibitor according to substrate and effector concentrations. However, the 3D structure for this enzyme is not yet reported. In this work, we characterized the NADP-ME2 allosteric site by structural modeling, molecular docking, normal mode analysis and mutagenesis. The regulatory site model and its docking analysis suggested that other C4 acids including malate, NADP-ME2 substrate, could also fit into fumarate’s pocket. Besides, a non-conserved cluster of hydrophobic residues in the second sphere of the allosteric site was identified. The substitution of one of those residues, L62, by a less flexible residue as tryptophan, resulted in a complete loss of fumarate activation and a reduction of substrate affinities for the active site. In addition, normal mode analysis indicated that conformational changes leading to the activation could originate in the region surrounding L62, extending through the allosteric site till the active site. Finally, the results in this work contribute to the understanding of structural determinants necessary for allosteric regulation providing new insights for enzyme optimization.
Graphic abstract








Similar content being viewed by others
References
Alvarez CE, Trajtenberg F, Larrieux N, Saigo M, Golic A, Andreo CS, Hogenhout SA, Mussi MA, Drincovich MF, Buschiazzo A (2018) The crystal structure of the malic enzyme from Candidatus Phytoplasma reveals the minimal structural determinants for a malic enzyme. Acta Crystallogr Sect D 74:332–340. https://doi.org/10.1107/S2059798318002759
Alvarez CE, Bovdilova A, Höppner A, Wolff CC, Saigo M, Trajtenberg F, Zhang T, Buschiazzo A, Nagel-Steger L, Drincovich MF, Lercher MJ, Maurino VG (2019) Molecular adaptations of NADP-malic enzyme for its function in C4 photosynthesis in grasses. Nat Plants 5:755–765. https://doi.org/10.1038/s41477-019-0451-7
Araújo WL, Nunes-Nesi A, Fernie AR (2011) Fumarate: multiple functions of a simple metabolite. Phytochemistry 72:838–843. https://doi.org/10.1016/j.phytochem.2011.02.028
Arias CL, Andreo CS, Drincovich MF, Gerrard Wheeler MC (2013) Fumarate and cytosolic pH as modulators of the synthesis or consumption of C(4) organic acids through NADP-malic enzyme in Arabidopsis thaliana. Plant Mol Biol 81:297–307. https://doi.org/10.1007/s11103-012-9999-6
Badia MB, Mans R, Lis AV, Tronconi MA, Arias CL, Maurino VG, Andreo CS, Drincovich MF, van Maris AJA, Gerrard Wheeler MC (2017) Specific Arabidopsis thaliana malic enzyme isoforms can provide anaplerotic pyruvate carboxylation function in Saccharomyces cerevisiae. FEBS J 284:654–665. https://doi.org/10.1111/febs.14013
Bakan A, Meireles LM, Bahar I (2011) ProDy: protein dynamics inferred from theory and experiments. Bioinformatics 27:1575–1577. https://doi.org/10.1093/bioinformatics/btr168
Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J (2000) The Protein Data Bank and the challenge of structural genomics. Nat Struct Biol 7:957–959. https://doi.org/10.1038/80734
Brooks B, Karplus M (1985) Normal modes for specific motions of macromolecules: application to the hinge-bending mode of lysozyme. Proc Natl Acad Sci USA 82:4995–4999. https://doi.org/10.1073/pnas.82.15.4995
Brown NJ, Palmer BG, Stanley S, Hajaji H, Janacek SH, Astley HM, Parsley K, Kajala K, Quick WP, Trenkamp S, Fernie AR, Maurini VG, Hibberd JM (2010) C4 acid decarboxylases required for C4 photosynthesis are active in the mid-vein of the C3 species Arabidopsis thaliana, and are important in sugar and amino acid metabolism. Plant J 61:122–133. https://doi.org/10.1111/j.1365-313X.2009.04040.x
Chang G, Tong L (2003) Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochemistry 42:12721–12733. https://doi.org/10.1021/bi035251
Chang H-C, Chen L-Y, Lu Y-H, Li M-Y, Chen Y-H, Lin C-H, Chang G-G (2007) Metal ions stabilize a dimeric molten globule state between the open and closed forms of malic enzyme. Biophys J 93:3977–3988. https://doi.org/10.1529/biophysj.107.111385
Coleman DE, Rao GSJ, Goldsmith EJ, Cook PF, Harris BG (2002) Crystal structure of the malic enzyme from Ascaris suum complexed with nicotinamide adenine dinucleotide at 2.3 Å resolution. Biochemistry 41:6928–6938. https://doi.org/10.1021/bi0255120
Detarsio E, Gerrard Wheeler MC, Campos Bermúdez VA, Andreo CS, Drincovich MF (2003) Maize C4 NADP-malic enzyme. Expression in Escherichia coli and characterization of site-directed mutants at the putative nucleoside-binding sites. J Biol Chem 278:13757–13764. https://doi.org/10.1074/jbc.M212530200
Detarsio E, Alvarez CE, Saigo M, Andreo CS, Drincovich MF (2007) Identification of domains involved in tetramerization and malate inhibition of maize C4-NADP-malic enzyme. J Biol Chem 282:6053–6060. https://doi.org/10.1074/jbc.M609436200
Dolinsky TJ, Nielsen JE, McCammon JA, Baker NA (2004) PDB2PQR: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res 32:665–667. https://doi.org/10.1093/nar/gkh381
Drincovich MF, Lara MV, Andreo CS, Maurino VG (2010) C4 decarboxylases: different solutions for the same biochemical problem, the provision of CO2 to rubisco in the bundle sheath cells. In: Raghavendra A, Sage R (eds) C4 photosynthesis and related CO2 concentrating mechanisms. Advances in photosynthesis and respiration. Springer, Dordrecht, pp 277–300
Eyal E, Lum G, Bahar I (2015) The anisotropic network model web server at 2015 (ANM 2.0). Bioinformatics 31:1487–1489. https://doi.org/10.1093/bioinformatics/btu847
Fernie AR, Martinoia E (2009) Malate. Jack of all trades or master of a few? Phytochemistry 70:828–832. https://doi.org/10.1016/j.phytochem.2009.04.023
Gerrard Wheeler MC, Tronconi MA, Drincovich MF, Andreo CS, Flügge UI, Maurino VG (2005) A comprehensive analysis of the NADP-malic enzyme gene family of Arabidopsis. Plant Physiol 139:39–51. https://doi.org/10.1104/pp.105.065953
Gerrard Wheeler MC, Arias CL, Tronconi MA, Maurino VG, Andreo CS, Drincovitch MF (2008) Arabidopsis thaliana NADP-malic enzyme isoforms: high degree of identity but clearly distinct properties. Plant Mol Biol 67:231–242. https://doi.org/10.1007/s11103-008-9313-9
Gerrard Wheeler MC, Arias CL, Maurino VG, Andreo CS, Drincovich MF (2009) Identification of domains involved in the allosteric regulation of cytosolic Arabidopsis thaliana NADP-malic enzymes. FEBS J 276:5665–5677. https://doi.org/10.1111/j.1742-4658.2009.07258.x
Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD (2006) Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics 22:2695–2696. https://doi.org/10.1093/bioinformatics/btl461
Hsieh J-Y, Chen S-H, Hung H-C (2009a) Functional roles of the tetramer organization of malic enzyme. J Biol Chem 284:18096–18105. https://doi.org/10.1074/jbc.M109.005082
Hsieh J-Y, Chiang Y-H, Chang K-Y, Hung H-C (2009b) Functional role of fumarate site Glu59 involved in allosteric regulation and subunit-subunit interaction of human mitochondrial NAD(P)+-dependent malic enzyme. FEBS J 276:983–994. https://doi.org/10.1111/j.1742-4658.2008.06834.x
Hsieh J-Y, Shih W-T, Kuo Y-H, Liu G-Y, Hung H-C (2019) Functional roles of metabolic intermediates in regulating the human mitochondrial NAD(P)+-dependent malic enzyme. Sci Rep 9:1–14. https://doi.org/10.1038/s41598-019-45282-0
Humphrey W, Dalk A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14(33–38):27–28. https://doi.org/10.1016/0263-7855(96)00018-5
Hung HC, Kuo MW, Chang GG, Liu GY (2005) Characterization of the functional role of allosteric site residue Asp 102 in the regulatory mechanism of human mitochondrial NAD(P)+-dependent malate dehydrogenase (malic enzyme). Biochem J 392:39–45. https://doi.org/10.1042/BJ20050641
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. https://doi.org/10.1107/s0021889892009944
Maurino VG, Engqvist MK (2015) 2-Hydroxy acids in plant metabolism. Arabidopsis Book 13:e0182. https://doi.org/10.1199/tab.0182
Monod J, Wyman J, Changeux J-P (1965) On the nature of allosteric transitions: a plausible model. J Mol Biol 12:88–118. https://doi.org/10.1016/S0022-2836(65)80285-6
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Murugan S, Hung H (2012) Biophysical characterization of the dimer and tetramer Interface interactions of the human cytosolic malic enzyme. PLoS ONE 7:1–11. https://doi.org/10.1371/journal.pone.0050143
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera - a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Rao GSJ, Coleman DE, Karsten WE, Cook PF, Harris BG (2003) Crystallographic studies on Ascaris suum NAD-malic enzyme bound to reduced cofactor and identification of an effector site. J Biol Chem 278:38051–38058. https://doi.org/10.1074/jbc.M305145200
Saigo M, Alvarez CE, Andreo CS, Drincovich MF (2013) Plastidial NADP-malic enzymes from grasses: unraveling the way to the C4 specific isoforms. Plant Physiol Biochem 63:39–48. https://doi.org/10.1016/j.plaphy.2012.11.009
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. https://doi.org/10.1006/jmbi.1993.1626
Tao X, Yang Z, Tong L (2003) Crystal structures of substrate complexes of malic enzyme and insights into the catalytic mechanism. Structure 11:1141–1150. https://doi.org/10.1016/S0969-2126(03)00168-0
Tronconi MA, Gerrard Wheeler MC, Martinatto A, Zubimendi JP, Andreo CS, Drincovich MF (2015) Allosteric substrate inhibition of Arabidopsis NAD-dependent malic enzyme 1 is released by fumarate. Phytochemistry 111:37–47. https://doi.org/10.1016/j.phytochem.2014.11.009
Tronconi MA, Andreo CS, Drincovich MF (2018) Chimeric structure of plant malic enzyme family: different evolutionary scenarios for NAD- and NADP-dependent isoforms. Front Plant Sci 9:1–15. https://doi.org/10.3389/fpls.2018.00565
Voll LM, Zell MB, Engelsdorf T, Saur A, Gerrard Wheeler MC, Drincovich MF, Weber APM, Maurino VG (2012) Loss of cytosolic NADP-malic enzyme 2 in Arabidopsis is associated with enhanced susceptibility towards Colletotrichum higginsianum. New Phytol 195:189–202. https://doi.org/10.1111/j.1469-8137.2012.04129.x
Xu Y, Bhargava G, Wu H, Loeber G, Tong L (1999) Crystal structure of human mitochondrial NAD ( P ) + -dependent malic enzyme : a new class of oxidative decarboxylases. Structure 7:877–889. https://doi.org/10.1016/S0969-2126(99)80115-4
Yang J, Zhang Y (2015) I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. https://doi.org/10.1093/nar/gkv342
Yang Z, Lanks CW, Tong L (2002a) Molecular mechanism for the regulation of human mitochondrial NAD(P)+-dependent malic enzyme by ATP and fumarate. Structure 10:951–960. https://doi.org/10.1016/S0969-2126(02)00788-8
Yang Z, Zhang H, Hung HC, Kuo CC, Tsai LC, Yuan HS, Chou WY, Chang GG (2002b) Structural studies of the pigeon cytosolic NADP(+) -dependent malic enzyme. Protein Sci 11:332–341. https://doi.org/10.1110/ps.38002
Acknowledgements
We thank Dra. Ana Bortolotti for her help with the use of the fluorometer and the IBR Institute for lending us this equipment. A MCGW, MFD and CEA belong to the Researcher Career of National Council of Scientific and Technical Research (CONICET); CLA participated as a fellow of the same institution. AMTS and CRR thanks Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ).
Funding
This work was supported by CONICET and National Agency for Promotion of Science and Technology. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) – Finance Code 001 (JFRM). This work was supported in part by a grant from Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Number E-26/203.179/2016.
Author information
Authors and Affiliations
Contributions
CEA conceived and led the project and together with MCGW and MFD designed and analyzed the experimental assays. CEA, CLA and MCGW designed the mutants and performed the kinetic and structural experiments. NCD performed and analyzed the NMA. JFRM and AMTS performed and analyzed the model structures and docking studies. CRR and AMTS supported all the computational work. All authors contributed to the writing of the manuscript and approved it.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1 (PNG 3015 KB)
Supplementary Figure S1. Sequence alignment of A. thaliana NADP-ME2 and HsmNAD(P)-ME. In light blue is shown the N-terminal residues modeled with I-TASSER; in red is highlighted the residues analyzed in this work and in pink are non-conservative substitution.
Supplementary file2 (PNG 1016 KB)
Supplementary Figure S2. ME2-L62W purification and initial characterization protocol. (a) SDS-PAGE of aliquots from His-trap purification stages: Escherichia coli protein extract (1), pellet (2) and soluble fraction (3) after centrifugation, binding (4), flow through (5), wash (6) and elute after enterokinase digestion (7). (b) Native PAGE comparing to NADP-ME2 (3-5 mU of each protein were loaded). Molecular weight markers were run in parallel in both gels (MW). (c) Elution profile by gel filtration chromatography. The elution volume corresponds to a native molecular mass of 277 kDa according to the calibration curve shown. (d) Circular dichroism spectra of mutant and parental proteins.
Supplementary file3 (AVI 7827 KB)
Video 1. 3D overview of NADP-ME2 twisting motion. In the video is shown the twisting motion and the region (11´´-residues 60-80, highlighted in red) proposed to be responsible of allosteric signal transmission.
Rights and permissions
About this article
Cite this article
Gerrard Wheeler, M.C., Arias, C.L., e Mello, J.F.R. et al. Structural insights into the allosteric site of Arabidopsis NADP-malic enzyme 2: role of the second sphere residues in the regulatory signal transmission. Plant Mol Biol 107, 37–48 (2021). https://doi.org/10.1007/s11103-021-01176-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-021-01176-2