Key message
PTR2 in Arabidopsis thaliana is negatively regulated by ABI4 and plays a key role in water uptake by seeds, ensuring that imbibed seeds proceed to germination.
Abstract
Peptide transporters (PTRs) transport nitrogen-containing substrates in a proton-dependent manner. Among the six PTRs in Arabidopsis thaliana, the physiological role of the tonoplast-localized, seed embryo abundant PTR2 is unknown. In the present study, a molecular physiological analysis of PTR2 was conducted using ptr2 mutants and PTR2CO complementation lines. Compared with the wild type, the ptr2 mutant showed ca. 6 h delay in testa rupture and consequently endosperm rupture because of 17% lower water content and 10% higher free abscisic acid (ABA) content. Constitutive overexpression of the PTR2 gene under the control of the Cauliflower mosaic virus (CaMV) 35S promoter in ptr2 mutants rescued the mutant phenotypes. After cold stratification, a transient increase in ABA INSENSITIVE4 (ABI4) transcript levels during induction of testa rupture was followed by a similar increase in PTR2 transcript levels, which peaked prior to endosperm rupture. The PTR2 promoter region containing multiple CCAC motifs was recognized by ABI4 in electrophoretic mobility shift assays, and PTR2 expression was repressed by 67% in ABI4 overexpression lines compared with the wild type, suggesting that PTR2 is an immediate downstream target of ABI4. Taken together, the results suggest that ABI4-dependent temporal regulation of PTR2 expression may influence water status during seed germination to promote the post-germinative growth of imbibed seeds.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Seeds comprise the seed coat, endosperm, and embryo. Germination of dormant seeds commences with the uptake of water and terminates with the emergence of the radicle through the seed coat (Bewley 1997). During germination, increase in the total water content of seeds follows a classic triphasic model, where the initial and final rapid water uptake periods (I and III, respectively) are intervened by the slow water uptake period II. Water imbibition-induced expansion of the embryo and endosperm leads to testa rupture (TR), which is followed by the protrusion of radicle through the endosperm, which leads to endosperm rupture (ER) (Weitbrecht et al. 2011).
Successful germination and post-germinative growth are partly determined by the endogenous plant hormones, including abscisic acid (ABA), gibberellic acid (GA), ethylene, and auxin, and the antagonism between GAs (which stimulate seed germination) and ABA (which establishes and maintains seed dormancy) is the primary factor that regulates seed germination (Pritchard et al. 2002; Seo et al. 2006). The endogenous level of ABA in dormant seeds declines upon imbibition during cold stratification (Chiwocha et al. 2005). Exogenously supplied ABA inhibits seed germination by affecting ER (Muller et al. 2006). ABA-mediated regulation of seed germination involves ABA receptor complexes consisting of PYRABACTIN RESISTANCE (PYR)/PYR1-LIKE (PYL)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR), type 2C protein phosphatases (PP2Cs), and SnRK2s (Raghavendra et al. 2010; Weiner et al. 2010). Multiple seed-abundant ABA-responsive transcription factors, including the B3 domain-containing ABA INSENSITIVE3 (ABI3), the APETALA2 (AP2) domain-containing ABI4, and the basic leucine zipper (bZIP) domain-containing ABI5 (Giraudat et al. 1992; Finkelstein et al. 1998; Finkelstein and Lynch 2000), operate in the ABA signaling pathways either in concert or independently. For instance, ABI4 inhibits ABA catabolism via the transcriptional repression of CYP707A (Shu et al. 2013) and catabolism of embryonic lipids (Penfield et al. 2006) during the germination process. MYB96 directly regulates ABI4 expression during embryonic lipid mobilization (Lee et al. 2015).
ABA is catabolized either by 8′-hydroxylation or glycosylation at the carboxyl group. Hydroxylation at the C-8′ of ABA is catalyzed by cytochrome P450-type mono-oxygenases (CYP707As) (Kushiro et al. 2004), and unstable 8'-hydroxy-ABA is then isomerized spontaneously to phaseic acid (Kepka et al. 2011). Glycosylation is catalyzed by eight ABA glycosyltransferases (GTs) in Arabidopsis thaliana (Lim et al. 2005). AtBG1 and AtBG2 inactivate ABA by converting it to ABA-glucose ester (ABA-GE) that accumulates in the vacuole or apoplast (Hartung et al. 2002; Lee et al. 2006). In Arabidopsis seeds, ABA metabolism and signaling-related genes are expressed in both endosperm and embryo, although the expression of ABI4 is higher in the embryo than in the endosperm (Penfield et al. 2006). Additionally, exogenous glucose (Glc) triggers ABA accumulation by activating the expression of ABA1 (encoding zeaxanthin epoxidase), ABA2 (encoding xanthoxin dehydrogenase), and ABA3 (encoding molybdenum cofactor sulfurase) genes, which in turn suppresses germination (Cheng et al. 2002; Price et al. 2003; Bossi et al. 2009).
Seed storage proteins are stored in the protein storage vacuole (or protein bodies), which is formed from vacuoles during seed maturation as a result of protein deposition and water displacement (Otegui et al. 2006). During seed imbibition and germination, storage proteins are hydrolyzed, and vacuoles fuse with each other, forming a central, lytic vacuole (Hunter et al. 2007; Zheng and Staehelin 2011). Once proteins are hydrolyzed, free amino acids and oligopeptides are transported to the cytosol by peptide transporters (PTRs), a type of symporter proteins, that cotransport protons (H+) and a wide range of nitrogen (N)-containing substrates, including nitrate, amino acids, and di-and tri-peptides (Chiang et al. 2004; Tsay et al. 2007), as well as GA, ABA, and jasmonates (Chiba et al. 2015). Among the six di- and tri-peptide transporting PTRs in Arabidopsis, PTR1 and PTR5 localize at the plasma membrane and perform distinct physiological functions; PTR1 regulates N uptake by the root, whereas PTR5 facilitates peptide transport to the germinating pollen (Komarova et al. 2008). PTR2 is highly expressed in the embryo (Rentsch et al. 1995; Song et al. 1996; Chiang et al. 2004; Léran et al. 2015) and endosperm (Dekkers et al. 2013), and localizes at the tonoplast (Komarova et al. 2012). Antisense suppression of PTR2 affects flowering and seed development but hardly affects seed germination (Song et al. 1997).
In the present study, we investigated the physiological function of PTR2 during early seed germination. The presence of multiple ABI4-binding motifs in the PTR2 promoter region led us to investigate the role of ABI4 in the regulation of PTR2 expression. The abundance of PTR2 transcripts in the endosperm (Dekkers et al. 2013) and embryo (Rentsch et al. 1995) during the early stage of seed germination (Supplementary Fig. S1), and localization of PTR2 at the tonoplast (Komarova et al. 2012) point to a role PTR2 in the regulation of the hydraulic status of germinating seeds. Indeed, the water content was lower in ptr2 mutant seeds and ABI4 negatively regulated PTR2 transcription during seed germination.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana ecotype Columbia (Col-0; WT), seven ptr mutants (ptr1–6) including ptr2-1 and ptr2-2 alleles, two PTR2 complementation lines (PTR2CO2 and PTR2CO5), abi4 mutant, and ABI4 overexpressor (ABI4OE) were used in this study. Seeds from seed batches grown and harvested at the same time and stored for less than 2 weeks were treated with 70% ethanol for 1 min and then sterilized with 0.8% sodium hypochlorite. After washing five times with sterilized distilled water (SDW), seeds were imbibed in water in the dark at 4 °C for 2 days to break seed dormancy and then transferred to continuous light (30 µmol m−2 s−1) at 22 °C. Seeds were sown in SDW supplemented with 2% Glc, 1 µM ABA, 50 µM GA, ABA inhibitors (20 µM nordihydroguaiaretic and 10 µM diniconazole), 20 mM glycine di-peptide (Gly-Gly), 20 mM leucine di-peptide (Leu-Leu), or 10 ∝g ml−1 of a transcription inhibitor, Cordycepin. The osmotic potential of SDW was adjusted to − 0.23, − 0.39, − 0.68, or − 0.91 Mpa with polyethylene glycol 4000 (PEG4000; Sigma-Aldrich).
Characterization of ptr mutants
T-DNA insertion mutant lines of all six Arabidopsis PTR genes were identified at The Arabidopsis Information Resource (TAIR) database (https://www.arabidopsis.org/index.jsp). Seeds of ptr1-1 (SLAK_131530), ptr2-1 (SALK_400_D08), ptr2-2 (SAL_65_B10), ptr3-2 (SALK_097591), ptr4-1 (SALK_062626), ptr5-2 (SALK_116120), and ptr6-1 (SALK_149283) were obtained from the Arabidopsis Biological Resource Center (ABRC) at Ohio State University, OH, USA. To map the T-DNA insertion sites, genomic DNA was isolated from the leaves of mutant plants using lysis buffer [200 mM Tris–HCl (pH 7.4), 250 mM NaCl, 25 mM EDTA (pH 8.0), and 0.5% sodium dodecyl sulfate (SDS)]. Reverse transcription PCR (RT-PCR) was carried out using the SALK LBa1, LBba1, and LB1 primers and PTR2 gene-specific primers (Supplementary Table S1). The resulting PCR products were sequenced and compared with the genomic sequence of each gene to map the T-DNA insertion site (Supplementary Fig. S2).
Generation of PTR2CO complementation lines
The coding sequence of PTR2 was amplified by PCR using sequence-specific primers (Supplementary Table S1). The PCR product was digested with XbaI and BamHI restriction endonucleases and cloned into the pJJ461 vector. The construct was confirmed by sequencing and transformed into Agrobacterium tumefaciens strain GV3101 by the freeze-thaw method. Transformation of ptr2 mutants was conducted using the floral dip method. PTR2CO lines with a single-copy of the hygromycin resistance (HygR) gene and PTR2 were identified by RT-PCR (Supplementary Fig. S2).
Extraction and analysis of seed storage proteins
Seed proteins was extracted using the chloroform-assisted phenol extraction (CAPE) method. Briefly, seeds (0.1 g fresh weight) were homogenized using a cold pestle and mortar in 1.0 ml buffer [0.25 M Tris–HCl (pH 7.5), 1% SDS, 14 mM dithiothreitol (DTT), and protease inhibitor cocktail (Roche)]. The homogenate was centrifuged at 12,000×g for 10 min. Then, 600 µl of the supernatant (embryo extract) was transferred to an Eppendorf tube, followed by the additional of an equal volume (600 µl) of chloroform. The mixture was mixed thoroughly by shaking for 10 min at room temperature. After the addition of 600 µl of buffered phenol (pH 8.0), the organic phase was transferred to two new 2.0 ml Eppendorf tubes (300 µl each). Then, five volumes of methanol containing 0.1 M ammonium acetate were added to each tube, and the mixture was incubated at − 20 °C for 1 h to precipitate the proteins. Proteins were separated by centrifugation at 15,000×g for 10 min and washed first with 100% cold acetone and then with 80% cold acetone. The protein pellet was air dried and resuspended in SDS sample buffer [50 mM Tris–HCl (pH 6.8), 100 mM DTT, 2% SDS, 0.1 bromophenol blue, and 10% glycerol]. Proteins were separated by SDS-polyacrylamide gel electrophoresis (PAGE) using 15% resolving and 5% stacking polyacrylamide gels.
Quantitative real-time PCR (qRT-PCR) analysis
Total RNA was extracted from dry and imbibed seeds using the Spectrum™ Plant Total RNA Kit (SIGMA) and treated with DNaseI (Takara). After NucleoSpin RNA Cleanup (Macherey-Nagel), cDNA was synthesized from 500 ng of total RNA using the iScript cDNA Synthesis Kit (Bio-Rad) and amplified by qRT-PCR on the CFX96 Real Time System (Bio-Rad) using the iQ™ SYBR Green Supermix (Bio-Rad), according to the manufacturer’s instructions. Reactions were performed in triplicate in a 10 µl volume, containing 5 µl SYBR Green Master Mix, 0.5 µl of each primer (10 pmol), 4 µl of 20-fold diluted cDNA, and 0.5 µl nuclease-free water, using the following conditions: 95 °C for 15 min, followed by 40 cycles of amplification at 94 °C for 10 s and 62 °C for 30 s, and lastly a final extension at 72 °C for 30 s. Fluorescence was measured at the end of each extension step. Amplification was followed by melting curve analysis, with continual fluorescence data acquisition during the 65 °C to 95 °C transition. A negative control containing water instead of cDNA template was included in each run. Raw data were analyzed with the CFX Manager software, and gene expression was normalized to the expression of AP2M (at5g46630) gene (Czechowski et al. 2005; Dekkers et al. 2012) to minimize variation in cDNA template levels. For each gene, a standard curve was generated using serial dilutions of cDNA, and the resultant PCR efficiency varied from 90 to 99.5%. To ensure that transcripts of single genes were amplified, qPCR amplicons were sequenced. Relative expression levels were calculated using the comparative threshold (Ct) cycle values, based on the 2−ΔΔCt method.
Quantification of seed ABA and water contents
Imbibed or germinating seeds (20 mg) were frozen in liquid N and ground using a pestle and mortar. The ground powder was immersed in 1 ml of 80% methanol and stored at 4 °C for 1 h. The extracts were filtered with C18 (Sep-Pak Vac) cartridges (Waters, USA) to remove pigments and other polar materials. The filtered solution was dried and concentrated using a rotary vacuum concentrator and suspended in Tris-buffered saline (TBS). The level of ABA was determined using the enzyme-linked immunosorbent assay (ELISA) Kit (Agdia). To measure the seed water content, ca. 330 seeds immersed in SDW were collected at a given time, and external liquid was removed using a silica-based membrane column by spinning at 12,000 rpm for 5 min. Then, dry seed weight was subtracted from the fresh seed weight to determine the amount of water absorbed by the seeds. External osmotic potential of varying concentrations of PEG4000 was calculated from the estimated osmolality (mol kg−1) (Money 1989).
Electrophoretic mobility shift assay (EMSA)
Glutathione S-transferase (GST)-tagged DNA-binding domain of ABI4 (GST-ABI4) was expressed in Escherichia coli and purified using a metal-affinity resin (Qiagen) (Finkelstein et al. 2011). Biotin-labeled PTR2 promoter fragments (− 360 to − 155 bp [P1]; − 760 to − 440 bp [P2]; − 1042 to − 739 bp [P3]; − 1775 to − 1583 bp [P4]) and competitor DNA fragments, including specific unlabeled fragment and ABI5 promoter fragment (− 915 to − 739 bp; positive control), were used with the purified GST-ABI4 protein in EMSAs (Chemiluminescent Light Shift EMSA kit; Pierce Biotechnology).
Measurement of GUS activity and transient gene expression
PTR2 and CaMV 35S promoters were cloned and fused to the β-glucuronidase (GUS) reporter gene in pCAMBIA3301 using primers listed in Supplementary Table S1. The resulting constructs were transiently transformed into the WT, abi4, and ABI4OE lines. Four-day-old seedlings grown in darkness were vacuum-infiltrated with Agrobacterium twice for 1 min each (Marion et al. 2008). The samples were incubated in the dark for 1 day and then grown under white light for 2 days. GUS activities were measured using a spectrofluorometer (LS-55; Perkin-Elmer) with the substrate 4-methylumbelliferyl-β-d-glucuronide (Sigma-Aldrich); 4-methylubelliferone (Sigma-Aldrich) was used for calibration. Protein content was determined using bovine serum albumin as the standard (Jaquinod et al. 2007). To perform GUS histochemical staining, germinating seeds were fixed by immersing in 90% (v/v) acetone. The seeds were then washed twice with a solution containing 50 mM sodium phosphate (pH 7), 0.5 mM K3Fe(CN)6, and 0.5 mM K4Fe(CN)6, and subsequently incubated in a staining solution containing 1 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Duchefa) (Lee et al. 2015).
Statistical analysis
Data (mean ± SE; n = 3–5) were analyzed using the two-tailed Student’s t test, after normality assessment. Means with p values less than 0.05 were considered statistically significant.
Results
Abundance of PTR2 transcripts, amino acids, and storage proteins in Arabidopsis seeds during germination
Publicly available microarray data show that transcript levels of PTR2 are highly abundant in both dry and germinating seeds, whereas those of the remaining five PTR genes are negligible (Supplementary Fig. S1A; Rentsch et al. 1995; Narsai et al. 2011; Dekkers et al. 2013, 2016). In this study, the transcript level of PTR2 in dry seeds was high and remained almost unchanged during cold stratification at 4 °C for 2 days but showed a transient increase during germination (Fig. 1a). This transient increase in PTR2 expression was inhibited by the transcription inhibitor cordycepin (Supplementary Fig. S1B). Thus, PTR2 activity appears to be regulated at the post-transcriptional level during early seed germination and then at the transcriptional level during late seed germination.
Seed germination kinetics in Arabidopsis. a Abundance of ABI4, PTR2, and CYP707A1/A2 transcripts in germinating Arabidopsis seeds. Transcript levels of each gene in germinating seeds were normalized to transcript levels in seeds immediately after stratification. b Changes in water, ABA, soluble protein (SP), and amino acid (aa) contents and rates of testa rupture (TR) and endosperm rupture (ER). Water, ABA, SP, and aa contents at a given time were normalized to those of dry seeds (ABA and SP) or post-germinated seeds (water and aa), respectively. ABA, 2.69 ± 0.14 pmol mg−1 seed DW; SP, 0.99 ± 0.01 mg mg−1 seed DW; aa, 0.68 ± 0.07 µmol mg−1 seed DW. The proportion of seeds with testa rupture (TR) or endosperm rupture (ER) was calculated using approximately 100 seeds at a given time. Images of germinating seeds at different stages, including no rupture, testa rupture (TR) with endosperm layer exposed, and endosperm rupture (ER) with radicle protruded, at various time points are provided on the top. Data represent mean ± standard error (SE; n = 3–4). Significant differences were detected at p < 0.05
The tonoplast-localized PTR2 would transport di- and tri-peptides, stored in protein bodies during seed development, to the cytosol. PTR2 would also transport small peptides generated from the degradation of seed storage proteins in embryo or cotyledons (Guerche et al. 1990; Pang et al. 1988) by proteases regulated at the post-transcriptional and/or post-translational level. If this is the case, a substantial amount of free amino acids and small peptides would be expected in dry and imbibed seeds, and a reverse change in free amino acid and storage protein contents would be expected when storage proteins undergo mobilization to support seed germination and subsequently seedling growth. The major seed storage proteins of Arabidopsis include the α- and β-subunits of the legumin-type 12S globulins (Pang et al. 1988) and the L- and S- subunits of 2S albumin (Guerche et al. 1990) (Fig. 1b). We observed that the contents of globulins and albumins stored in seeds steadily decreased, whereas those of free amino acids increased shortly after stratification but ahead of TR, as expected. However, the inverse change between proteins and amino acids was not observed; instead, the protein and amino acid contents remained unchanged during stratification.
We also quantified the water and ABA contents of germinating seeds, as aquaporin-independent uptake of water (Van der Willigen et al. 2006; Obroucheva 2012) and reduction of ABA content (Chiwocha et al. 2005; Muller et al. 2006) are major events during stratification. During water uptake phase II under low temperature, the ABA content of seeds decreased to the lowest level 1 day prior to the start of the decline in seed storage protein content, and then remained almost unchanged until ER. This decrease in the ABA level is mostly due to the transcriptional activation of CYP707A1 and CYP707A2 (Fig. 1a; Dekkers et al. 2013). These changes in CYP707A1 and CYP707A2 transcript levels after stratification were cordycepin-sensitive (Supplementary Fig. S1B), suggesting a transcriptional regulation of both these genes during seed germination. However, transcripts of CYP707A3, CYP707A4, GTs, and BGs showed minimal or no changes (data not shown).
Mutation of PTR2 delays TR
To identify the physiological role of PTR2 in the seed germination process, ptr2 mutants were treated with Glc, ABA, and GA either alone or in combination. The germination of both WT seeds started at 12 h after stratification (HAS) and was almost complete at 36 HAS (Figs. 1, 2). However, ptr2 mutant seeds displayed an approximately 6 h delay mainly because of delayed TR (Fig. 2a), and consequently delayed ER, albeit to a lower extent (Fig. 2b). This delayed TR phenotype of the ptr2 mutant could not be recovered by the exogenous application of GA (Supplementary Fig. S3A) or amplified by that of ABA and Glc (Fig. 2a, b). Additionally, the delay in TR was similar between the allelic mutants, ptr2-1 and ptr2-2 (Supplementary Fig. S3B), and was fully rescued by the overexpression of PTR2 under the control of the CaMV 35S promoter (35SCaMVp::PTR2) in the complementation lines PTR2CO2 and PTR2CO5 (Fig. 2c, d). Furthermore, absence of TR delay in other ptr mutants strongly suggests a unique role of PTR2 in seed germination (Supplementary Fig. S3B).
Delay in testa rupture (TR) and endosperm rupture (ER) in ptr2 mutant seeds. a, b Additive effects of ABA and glucose (Glc) on the germination of ptr2 mutant seeds. Col-0 (wild type; WT) and ptr2-1 mutant seeds were germinated on sterile distilled water (SDW; Con), 2% Glc, 1 µM ABA, or 2% Glc + 1 µM ABA. c, d Complementation of the germination delay phenotype of ptr2-1 mutant seeds. Seeds of Col-0 (WT), ptr2-1, and PTR2CO2 and PTR2CO5 complementation lines were germinated on SDW. Data represent mean ± SE (n = 3–4)
Reduction in the water content of ptr2 mutant seeds during germination
In Arabidopsis, swelling of the embryo and endosperm due to imbibition leads to the mobilization of storage reserves and translation of de novo synthesized or stored mRNAs (Weitbrecht et al. 2011), among other complex multiple events, ultimately leading to TR. Therefore, we determined the water content of germinating ptr2-1 seeds exhibiting the delayed TR phenotype. Rapid water uptake during phase I was not affected by the PTR2 mutation. However, water uptake during phase II and phase III was lowered by 17% and 8%, respectively (Fig. 3a). This phenotype was rescued by the expression of 35SCaMVp::PTR2 in ptr2-1 mutant (Fig. 3b), confirming that PTR2 is at least in part involved in water uptake during phases II and III. Osmotic potential contributed by PTR2 was estimated at − 0.21 MPa, based on the time required to reach 50% TR or ER (Fig. 3c).
Role of PTR2 in water uptake during seed germination. a Changes in water content were analyzed during the progression of seed germination. Col-0 (WT) and ptr2-1 mutant seeds were germinated on SDW (Con). Fractional difference is the difference in water content at a given time point between WT and ptr2-1 mutant seeds. Data represent mean ± SE (n = 3–4).b Complementation of the water deficit phenotype of the ptr2 mutant. Seeds of Col-0 (WT), ptr2-1 mutant, andPTR2CO2 complementation line were used 12 h after stratification. Data represent mean ± SE (n = 3–4). Statistically significant differences are indicated with an asterisk (*p < 0.05).c Estimation of the water potential of the ptr2 mutant during seed germination. Water potential was calculated based on the linear positive correlation between the water potential generated by PEG4000 and time needed to reach 50% TR (r2 = 0.992) or ER (r2 = 0.995). Data represent mean ± SE (n = 3–4)
Increase in ABA content in ptr2 mutant seeds during the early phase of germination
A linear interaction between ABA contents and water potential during seed germination (Welbaum et al. 1990) implies that ptr2 mutant seeds with lowered water content should contain more ABA than WT seeds at the same germination stage. Indeed, the ABA content of germinating ptr2 seeds was 10–16% higher than that of WT seeds as well as those of PTR2CO2 and PTR2CO5. Furthermore, PEG4000 treated WT, ptr2-1, and PTR2CO seeds showed ca. 60% higher ABA contents than untreated seeds (Table 1). Increase in the ABA content of WT seeds caused by PTR2 mutation and PEG4000 treatment was comparable to that caused by Glc and diniconazole treatments, respectively. Thus, the PTR2 mutation seems to impair ABA catabolism during seed germination to a similar extent as that caused by exogenous Glc application.
ABI4 acts as a repressor of PTR2
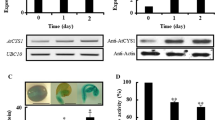
The PTR2 promoter contains multiple ABI4-binding motifs of the sequence CCAC (Bossi et al. 2009; Reeves et al. 2011; Fig. 4a). To test whether these CCAC sequences are recognized by ABI4, we conducted EMSAs using four CCAC motif-containing PTR2 promoter fragments (P1–4) and the GST-ABI4 fusion protein. The recombinant ABI4 protein containing only the DNA-binding domain bound directly to all PTR2 promoter fragments, like ABI5 (a positive control), but this binding was inhibited by unlabeled, cold fragments (Fig. 4a). If ABI4 acts immediately upstream of PTR2, then the transcript level of PTR2 should be affected in abi4 as well as in ABI4OE plants but in an opposite manner. Compared with the WT, ABI4OE plants showed a significantly lower transcript level of PTR2, but the abi4 mutant failed to show a significant enhancement in PTR2 transcript level (Fig. 4b). This inhibitory effect was also conserved in seeds treated with Glc (Fig. 4b), where endogenous ABA level was enhanced (Table 1). Taken together, these data strongly suggest that PTR2 transcription is negatively regulated by ABI4. To confirm these results in vivo, PTR2 promoter was fused to the GUS reporter gene (PTR2p::GUS) and transformed into hypocotyls of WT, abi4, and ABI4OE. The expression of PTR2-GUS was lowered by 10% in the ABI4OE background, consistent with the results of the EMSA, thus confirming that ABI4 represses PTR2 transcription by directly binding to the CCAC motifs in PTR2 promoter (Fig. 4c).
Repression of PTR2 transcription by ABI4. a Schematic representation of the PTR2 promoter containing six CCAC motifs. Four promoter fragments (P1, − 360 to − 155 bp; P2, − 760 to − 440 bp; P3, − 1042 to − 739 bp; P4, − 1775 to − 1583 bp), each containing a single CCAC motif (red square), were used for the electrophoretic mobility shift assay (EMSA). Arrows indicate primers used for cloning. PTR2 promoter fragments (P1–P4) were co-incubated with purified GST-ABI4 (lanes 1–11) and GST-ABI5 (lanes 13 and 14) DNA-binding domain protein, and GST (lane 12). Biotin-labeled DNA fragments (30 fmol) were incubated for 30 min at 22 °C with (lanes 2, 4, 6, 8, 9, 10, 11, 12, and 14) or without (lanes 1, 3, 5, 7, and 13) purified GST-ABI4 (6 μg; lanes 1 to 6, 2 μg; lane 8, 4 μg; lane 9, 8 μg; lanes 10 and 11), GST-ABI5 (6 μg; lane 14; control) or GST (3 μg; lane 12; control). A 200-fold molar excess of the unlabeled P4 inhibitor was included (lane 11). b Quantitative real-time PCR (qRT-PCR) analysis of PTR2 expression in Col-0, abi4, and ABI4OE seeds at 12 h after stratification. Seeds were germinated on SDW (Con) or 2% Glc. Data represent mean ± SE (n = 3–4). Statistically significant differences are indicated with an asterisk (*p < 0.05). c Transient expression of PTR2 promoter-driven GUS gene (PTR2p::GUS) in abi4 and ABI4OE. GUS activity (nmol min−1 μg protein−1) was measured using the substrate 4-methylumbelliferyl-β-d-glucuronide. Data represent mean ± SE (n = 3–4). Statistically significant differences are indicated with an asterisk (*p < 0.05)
Discussion
Unlike endospermless species, Arabidopsis exhibits two consecutive germination processes, including TR and ER (Muller et al. 2006). The PTR2 protein is abundant in the embryo (Rentsch et al. 1995; Supplementary Fig. S4) and endosperm (Dekkers et al. 2013). Although mutation of the PTR2 gene did not affect the canonical triphasic water uptake kinetics, it resulted in lower seed water content, especially during phase II and early phase III. The dependence of TR, and hence ER, on external water potential (Fig. 3c) implies the importance of the endogenous osmotic potential in determining the expansion of the embryo and endosperm tissues. Currently, it is unknown how the biochemical loss of PTR2 is involved in the water-driven seed swelling process. Similar to amino acids, which act as endogenous osmotic solutes like sugars and potassium ions (Bove et al. 2001), di- and tri-peptides seem to also act as endogenous osmotica. Then, increases in the cytosolic level of small peptides via transcriptionally or post-transcriptionally activated PTR2 would increase the cytosolic osmotic potential (to approximately − 0.21 Mpa, as a proxy measure), contributing to aquaporin-independent water diffusion into cells (Fig. 5). Expression of PTR2p::GUS in the germinating embryo during phase II (Supplementary Fig. S4), which is shaped by ABI4, suggests that PTR2 is involved in water uptake rather than in N mobilization from the endosperm to the growing axis. Our results are consistent with those of Song et al. (1997); the authors showed that ptr2 mutant alleles exhibited comparable seedling growth as the antisense mutants.
Schematic representation of the role of PTR2 in water uptake during early seed germination in Arabidopsis. In endosperm and embryo tissues, PTR2 is activated at the post-transcriptional level, whilst its transcriptional regulation confines mostly in the embryo due to the presence of its negative regulator ABI4. Once activated and localized to the vacuolar membrane, PTR2 transports small peptides stored during seed maturation or generated by the enzymatic hydrolysis of seed storage proteins to the cytosol. Water potential generated by the accumulation of small peptides in the cytosol facilitates water uptake by diffusion, which leads to TR and ER in a sequential manner. Arrow and ⊥ indicate activation and repression, respectively
Di- and tri-peptide sources available for the tonoplast-localized PTR2 seem different between the first half and second half of the water uptake phase II. Time-course analysis of water and storage protein contents of seeds clearly revealed that the inhibition of water uptake observed in the ptr2 mutant seeds during stratification occurred earlier than the decline in the storage protein content. Thus, PTR2 in seeds seems to initially transport di- and tri-peptides stored in protein storage vacuole or protein bodies to the cytosol, followed by the peptides hydrolyzed from storage proteins. Accordingly, amino acids and small peptides that accumulate in the cytosol would serve primarily as osmotica, contributing to the swelling of the embryo and endosperm, a prerequisite for TR. At the later stage of seed germination and subsequent seedling growth, these amino acids and small peptides would be utilized for further N metabolism. The content and composition of seed storage proteins were comparable among the dry seeds of the WT, ptr2-1 mutant, and PTR2OEs matured at both 22 °C and 16 °C (Supplementary Fig. S5), thus ruling out the role of PTR2 in the mobilization of seed storage proteins during seed maturation.
PTR2 belongs to NRT1/PTR family (NPF) proteins. In addition to nitrate and di- and tri-peptides, some NPFs are involved in the uptake of other substrates, such as nitrite, chloride, glucosinolates, auxin, ABA, GA, and jasmonates (see review Corratge-Faillie and Lacombe 2017). Like PTR1 and PTR5 (Chiba et al. 2015), PTR2 might use different substrates, such as ABA and jasmonates. In the present study, we did not find any evidence that water uptake was influenced by these substrates, including IAA, GA, ABA, and glucose. This rules out the possibility that the transport activity of other substrates is involved in the observed phenotype, namely, water uptake driven seed germination. It is also unlikely that the post-germination process would be controlled by PTR2 transport activity, considering that the mutant phenotype was not observed during post-germination or afterwards during the growth and development stages.
PTR2 transcripts (Rentsch et al. 1995; Dekkers et al. 2013; Supplementary Fig. S4) and 12S globulins (Pang et al. 1988) are stored in both the endosperm and embryo during seed maturation. Furthermore, a transient increase in PTR2 expression during the transition from TR to ER is sensitive to cordycepin, a transcription inhibitor. Thus, PTR2 in germinating seeds is likely activated either post-transcriptionally or transcriptionally. While post-transcriptional activation of PTR2 could occur in both seed tissues, re-repression of ABI4-dependent transcriptional suppression of PTR2 could occur only in the embryo (Fig. 5) since ABI4 is mostly active in the embryo, unlike ABI3 and ABI5, which function in both the embryo and endosperm during late phase II and early phase III. Accordingly, ABI4 seems to act as a spatiotemporal regulator of PTR2 expression necessary for water uptake-dependent ER. The expression of ABI4 is induced by high endogenous ABA levels during seed dormancy or exogenously supplied ABA, leading to the inhibition of ER. Thus, transient regulation of ABI4 during the TR induction phase, when ABA level is low, implies the presence of an upstream regulator of ABI4. The DELLA protein RGL2 and the MYB protein MYB96 (Penfield et al. 2005, 2006; Lee et al. 2015; see review Gazzarrini and Tsai 2015) might act as putative regulators of ABI4, although this needs to be confirmed in a future study. Additionally, the redundant regulation of PTR2 by ABI3 and ABI4 (Giraudat et al. 1992; Finkelstein et al. 1998; Finkelstein and Lynch 2000) should be also tested.
Taken together, our results suggest that disruption of PTR2 function results in lower water content in the endosperm and embryo during early seed germination in Arabidopsis. Tonoplast-localized PTR2 is likely activated post-transcriptionally upon water imbibition prior to TR and transcriptionally by ABI4 prior to onset of ER. In the expanding embryo, ABI4 may directly repress PTR2 expression by binding to CCAC motifs in its promoter region. Further work will be required to demonstrate directly the osmotic potential build-up by PTR2-dependent increases in cytosolic di- and tri-peptides in germinating seeds.
Abbreviations
- ABA:
-
Abscisic acid
- ABI:
-
ABA insensitive
- ER:
-
Endosperm rupture
- Glc:
-
Glucose
- PTR:
-
Peptide transporter
- TFs:
-
Transcription factors
- TR:
-
Testa rupture
- WT:
-
Wild type
References
Bewley JD (1997) Seed germination and dormancy. Plant Cell 9:1055–1066
Bossi F, Cordoba E, Dupré P, Mendoza MS, Román CS, León P (2009) The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. Plant J 59:359–374
Bove J, Jullien M, Grappin P (2001) Functional genomics in the study of seed germination. Genome Biol 3:1002.1-1002.5
Cheng WH, Endo A, Zhou L, Penney J, Chen HC, Arroyo A, Leon P, Nambara E, Asami T, Seo M (2002) A unique short-chain dehydrogenase/reductase in Arabidopsis glucose signaling and abscisic acid biosynthesis and functions. Plant Cell 14:2723–2743
Chiba Y, Shimizu T, Miyakawa S, Kanno Y, Koshiba T, Kamiya Y, Seo M (2015) Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J Plant Res 128:679–686
Chiang C-S, Stacey G, Tsay Y-F (2004) Mechanisms and functional properties of two peptide transporters, AtPTR2 and fPTR2. J Biol Chem 279:30150–30157
Chiwocha SDS, Cutler AJ, Abrams SR, Ambrose SJ, Yang J, Ross ARS, Kermode AR (2005) The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J 42:35–48
Corratge-Faillie C, Lacombe B (2017) Substate (un)specificity of Arabidopsis NRT1/PRT Family (NPF) proteins. J Exp Bot 68:3107–3113
Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR (2005) Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol 139:5–17
Dekkers BJ, Pearce S, van Bolderen-Veldkamp RP, Holdsworth MJ, Bentsink L (2016) Dormant and after-Ripened Arabidopsis thaliana seeds are distinguished by early transcriptional differences in the imbibed state. Front Plant Sci 7:1323–1337
Dekkers BJ, Pearce S, van Bolderen-Veldkamp RP, Marshall A, Widera P, Drost HG, Bassel GW, Muller K, King JR, Wood AT, Grosse I, Quint M, Krasnogor N, Leubner-Metzger G, Holdsworth MJ, Bentsink L (2013) Transcriptional dynamics of two seed compartments with opposing roles in Arabidopsis seed germination. Plant Physiol 163:205–215
Dekkers BJ, Willems L, Bassel GW, van Bolderen-Veldkamp RP, Ligterink W, Hilhorst HW, Bentsink L (2012) Identification of reference genes for RT-qPCR expression analysis in Arabidopsis and tomato seeds. Plant Cell Physiol 53:28–37
Finkelstein R, Lynch T (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12:599–609
Finkelstein R, Lynch T, Reeves W, Petitfils M, Mostachetti M (2011) Accumulation of the transcription factor ABA-insensitive (ABI)4 is tightly regulated post-transcriptionally. J Exp Bot 62:3971–3979
Finkelstein R, Wang ML, Lynch TJ, Rao S, Goodman HM (1998) The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. Plant Cell 10:1043–1054
Gazzarrini S, Tsai A (2015) Hormone cross-talk during seed germination. Essays Biochem 58:151–164
Giraudat J, Hauge BM, Valon C, Smalle J, Parcy F, Goodman HM (1992) Isolation of the Arabidopsis ABI3 gene by positional cloning. Plant Cell 4:1251–1261
Guerche P, Tire C, De Sa FG, De Clercq A, Van Montagu M, Krebbers E (1990) Differential expression of the Arabidopsis 2S albumin genes and the effect of increasing gene family size. Plant Cell 2:469–478
Hartung W, Sauter A, Hose E (2002) Abscisic acid in the xylem: where does it come from, where does it go to? J Exp Bot 53:27–32
Hunter PR, Craddock CP, Di Benedetto S, Roberts LM, Frigerio L (2007) Fluorescent reporter proteins for the tonoplast and the vacuolar lumen identify a single vacuolar compartment in Arabidopsis cells. Plant Physiol 145:1371–1382
Jaquinod M, Villiers F, Kieffer-Jaquinod S, Hugouvieux V, Bruley C, Garin J, Bourguignon J (2007) A proteomics dissection of Arabidopsis thaliana vacuoles isolated from cell culture. Mol Cell Proteom 6:394–412
Kepka M, Benson CL, Gonugunta VK, Nelson KM, Christmann A, Grill E, Abrams SR (2011) Action of natural abscisic acid precursors and catabolites on abscisic acid receptor complexes. Plant Physiol 157:2108–2119
Komarova NY, Meier S, Meier A, Grotemeyer MS, Rentsch D (2012) Determinants for Arabidopsis peptide transporter targeting to the tonoplast or plasma membrane. Traffic 13:1090–1105
Komarova NY, Thor K, Gubler A, Meier S, Dietrich D, Weichert A, Suter Grotemeyer M, Tegeder M, Rentsch D (2008) AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiol 148:856–869
Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'-hydroxylases: key enzymes in ABA catabolism. EMBO J 23:1647–1656
Lee KH, Piao HL, Kim HY, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee IJ, Hwang I (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126:1109–1120
Lee K, Lee HG, Yoon S, Kim HU, Seo PJ (2015) The Arabidopsis MYB96 transcription factor is a positive regulator of ABI4 in the control of seed germination. Plant Physiol 168:677–689
Léran S, Garg B, Boursiac Y, Corratgé-Faillie C, Brachet C, Tillard P, Gojon A, Lacombe B (2015) AtNPF5.5, a nitrate transporter affecting nitrogen accumulation in Arabidopsis embryo. Sci Rep 5:7962
Lim EK, Doucet CJ, Hou B, Jackson RG, Abrams SR, Bowles DJ (2005) Resolution of (+)-abscisic acid using an Arabidopsis glycosyltransferase. Tetrahedron-Asymmetry 16:143–147
Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD (2008) Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. Plant J 56:169–179
Money NP (1989) Osmotic pressure of aqueous polyethylene glycols. Plant Physiol 91:766–769
Muller K, Tintelnot S, Leubner-Metzger G (2006) Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryo induced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol 47:864–877
Narsai R, Law SR, Carrie C, Xu L, Whelan J (2011) In-depth temporal transcriptome profiling reveals a crucial developmental switch with roles for RNA processing and organelle metabolism that are essential for germination in Arabidopsis. Plant Physiol 157:1342–1362
Obroucheva NV (2012) Transition from hormonal to non-hormonal regulation as exemplified by seed dormancy release and germination triggering. Russ J Plant Physiol 59:591–600
Otegui MS, Herder R, Schulze J, Jung R, Staehelin LA (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18:2567–2581
Pang PP, Pruitt RE, Meyerowitz EM (1988) Molecular cloning, genomic organization, expression and evolution of 12S seed storage protein genes of Arabidopsis thaliana. Plant Mol Biol 11:805–820
Penfield S, Graham S, Graham IA (2005) Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem Soc Trans 33:380–383
Penfield S, Li Y, Gilday AD, Graham S, Graham IA (2006) Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18:1887–1899
Price J, Li TC, Kang SG, Na JK, Jang JC (2003) Mechanisms of Glc-signaling during germination of Arabidopsis. Plant Physiol 132:1424–1438
Pritchard SL, Charlton WL, Baker A, Graham IA (2002) Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J 31:639–647
Raghavendra AS, Gonugunta VK, Christmann A, Grill E (2010) ABA perception and signalling. Trends Plant Sci 15:395–401
Reeves WM, Lynch TJ, Mobin R, Finkelstein RR (2011) Direct targets of the transcription factors ABA-Insensitive (ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol Biol 75:347–363
Rentsch D, Laloi M, Rouhara I, Schmelzer E, Delrot S, Frommer WB (1995) NTR1 encodes a high affinity oligopeptide transporter in Arabidopsis. FEBS Lett 370:264–268
Seo M, Hanada A, Kuwahara A, Endo A, Okamoto M, Yamauchi Y, North H, Marion-Poll A, Sun TP, Koshiba T, Kamiya Y, Yamaguchi S, Nambara E (2006) Regulation of hormone metabolism in Arabidopsis seeds: phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J 48:354–366
Shu K, Zhang H, Wang S, Chen M, Wu Y, Tang S, Liu C, Feng Y, Cao X, Xie Q (2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis PLoS Genet 9:E1003577
Song W, Koh S, Czako M, Marton L, Drenkard E, Becker JM, Stacey G (1997) Antisense expression of the peptide transport gene AtPTR2-B delays flowering and arrests seed development in transgenic Arabidopsis plants. Plant Physiol 114:927–935
Song W, Steiner HY, Zhang L, Naider F, Stacey G, Becker JM (1996) Cloning of a second Arabidopsis peptide transport gene. Plant Physiol 110:171–178
Tsay YF, Chiu CC, Tsai CB, Ho CH, Hsu PK (2007) Nitrate transporters and peptide transporters. FEBS Lett 581:2290–2300
Vander Willigen C, Postaire O, Tournaire-Roux C, Boursiac Y, Maurel C (2006) Expression and inhibition of aquaporins in germinating Arabidopsis seeds. Plant Cell Physiol 47:1241–1250
Weiner JJ, Peterson FC, Volkman BF, Cutler SR (2010) Structural and functional insights into core ABA signaling. Curr Opin Plant Biol 13:495–502
Weitbrecht K, Muller K, Leubner-Metzger G (2011) First off the mark: early seed germination. J Exp Bot 62:3289–3309
Welbaum GE, Tissaoui T, Bradford KJ (1990) Water relations of seed development and germination in muskmelon (Cucumis melo L.). Plant Physiol 92:1029–1037
Zheng H, Staehelin LA (2011) Protein storage vacuoles are transformed into lytic vacuoles in root meristematic cells of germinating seedlings by multiple, cell type-specific mechanisms. Plant Physiol 155:2023–2035
Acknowledgements
This work was funded by the Cooperative Research Program for Agriculture Science & Technology Development (PJ01279203), RDA and the Collaborative Genome Program of the Korea Institute of Marine Science and Technology Promotion (KIMST) funded by the Ministry of Oceans and Fisheries (MOF) (No. 20180430), Republic of Korea.
Author information
Authors and Affiliations
Contributions
Y-IP and C-SK designed the research; M-GC, EJK, and J-YS performed the experiments; S-BC, S-WC, CSP, C-SK, and Y-IP wrote the manuscript. All authors revised and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Choi, MG., Kim, E.J., Song, JY. et al. Peptide transporter2 (PTR2) enhances water uptake during early seed germination in Arabidopsis thaliana. Plant Mol Biol 102, 615–624 (2020). https://doi.org/10.1007/s11103-020-00967-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-020-00967-3