Abstract
Key message
Metabolic module, gene expression pattern and PLS modeling were integrated to precisely identify the terpene synthase responsible for sesquiterpene formation. Functional characterization confirmed the feasibility and sensitivity of this strategy.
Abstract
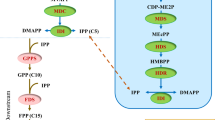
Plant secondary metabolite biosynthetic pathway elucidation is crucial for the production of these compounds with metabolic engineering. In this study, an integrated strategy was employed to predict the gene function of sesquiterpene synthase (STS) genes using turmeric as a model. Parallel analysis of gene expression patterns and metabolite modules narrowed the candidates into an STS group in which the STSs showed a similar expression pattern. The projections to latent structures by means of partial least squares model was further employed to establish a clear relationship between the candidate STS genes and metabolites and to predict three STSs (ClTPS16, ClTPS15 and ClTPS14) involved in the biosynthesis of several sesquiterpene skeletons. Functional characterization revealed that zingiberene and β-sesquiphellandrene were the major products of ClTPS16, and β-eudesmol was produced by ClTPS15, both of which indicated the accuracy of the prediction. Functional characterization of a control STS, ClTPS1, produced a small amount of β-sesquiphellandrene, as predicted, which confirmed the sensitivity of metabolite module analysis. This integrated strategy provides a methodology for gene function predictions, which represents a substantial improvement in the elucidation of biosynthetic pathways in nonmodel plants.





Similar content being viewed by others
Abbreviations
- DGE:
-
Digital gene expression profiling
- FPP:
-
Farnesyl pyrophosphate
- HCA:
-
Hierarchical cluster analysis
- NIST:
-
National Institute of Standards and Technology
- MET-IDEA:
-
Metabolomics ion-based data extraction algorithm
- PLS:
-
Projections to latent structures by means of partial least squares
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction analysis
- STS:
-
Sesquiterpene synthase
- SD-Trp:
-
Synthetic defined medium lacking tryptophan
- TPS:
-
Terpene synthase
References
Acharjee A, Kloosterman B, de Vos RC, Werij JS, Bachem CW, Visser RG, Maliepaard C (2011) Data integration and network reconstruction with omics data using random forest regression in potato. Anal Chim Acta 705:56–63. https://doi.org/10.1016/j.aca.2011.03.050
Ajikumar PK, Xiao WH, Tyo KE, Wang Y, Simeon F, Leonard E, Mucha O, Phon TH, Pfeifer B, Stephanopoulos G (2010) Isoprenoid pathway optimization for Taxol precursor overproduction in Escherichia coli. Science 330:70–74. https://doi.org/10.1126/science.1191652
Banerjee A, Hamberger B (2018) P450s controlling metabolic bifurcations in plant terpene specialized metabolism. Phytochem Rev 17:81–111. https://doi.org/10.1007/s11101-017-9530-4
Bino RJ, Hall RD, Fiehn O, Kopka J, Saito K, Draper J, Nikolau BJ, Mendes P, Roessner-Tunali U, Beale MH, Trethewey RN, Lange BM, Wurtele ES, Sumner LW (2004) Potential of metabolomics as a functional genomics tool. Trends Plant Sci 9:418–425. https://doi.org/10.1016/j.tplants.2004.07.004
Bleeker PM, Spyropoulou EA, Diergaarde PJ, Volpin H, DeBoth MTJ, Zerbe P, Bohlmann J, Falara V, Matsuba Y, Pichersky E, Haring MA, Schuurink RC (2011) RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Mol Biol 77:323. https://doi.org/10.1007/s11103-011-9813-x
Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95:4126–4133. https://doi.org/10.2307/44860
Bouhaddani SE, Houwing-Duistermaat J, Salo P, Perola M, Jongbloed G, Uh HW (2016) Evaluation of O2PLS in omics data integration. BMC Bioinform 17:11. https://doi.org/10.1186/s12859-015-0854-z
Braga ME, Leal PF, Carvalho JE, Meireles MA (2003) Comparison of yield, composition, and antioxidant activity of turmeric (Curcuma longa L.) extracts obtained using various techniques. J Agric Food Chem 51:6604–6611. https://doi.org/10.1021/jf0345550
Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW (2006) MET-IDEA: data extraction tool for mass spectrometry-based metabolomics. Anal Chem 78:4334–4341. https://doi.org/10.1021/ac0521596
Cavill R, Jennen D, Kleinjans J, Briede JJ (2016) Transcriptomic and metabolomic data integration. Brief Bioinform 17:891–901. https://doi.org/10.1093/bib/bbv090
Chen X, Facchini PJ (2014) Short-chain dehydrogenase/reductase catalyzing the final step of noscapine biosynthesis is localized to laticifers in opium poppy. Plant J 77:173–184. https://doi.org/10.1111/tpj.12379
Copolovici L, Kännaste A, Remmel T, Niinemets Ü (2014) Volatile organic compound emissions from Alnus glutinosa under interacting drought and herbivory stresses. Environ Exp Bot 100:55–63. https://doi.org/10.1016/j.envexpbot.2013.12.011
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstr C, Wold S (2006a) Multi-and megavariate data analysis part I: basic principles and applications. Second revised and enlarged edition. Umetrics Academy, Sweden, p 64
Eriksson L, Johansson E, Kettaneh-Wold N, Trygg J, Wikstr C, Wold S (2006b) Multi-and megavariate data analysis part I: basic principles and applications, second revised and enlarged edition. Umetrics Academy Umetrics, Sweden, p 208
Galanie S, Thodey K, Trenchard IJ, Filsinger Interrante M, Smolke CD (2015) Complete biosynthesis of opioids in yeast. Science 349:1095–1100. https://doi.org/10.1126/science.aac9373
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, Chen Z, Mauceli E, Hacohen N, Gnirke A, Rhind N, di Palma F, Birren BW, Nusbaum C, Lindblad-Toh K, Friedman N, Regev A (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Hirai MY, Yano M, Goodenowe DB, Kanaya S, Kimura T, Awazuhara M, Arita M, Fujiwara T, Saito K (2004) Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc Natl Acad Sci USA 101:10205–10210. https://doi.org/10.1073/pnas.0403218101
Jiang H, Xie Z, Koo HJ, McLaughlin SP, Timmermann BN, Gang DR (2006) Metabolic profiling and phylogenetic analysis of medicinal Zingiber species: tools for authentication of ginger (Zingiber officinale Rosc). Phytochemistry 67:1673–1685. https://doi.org/10.1016/j.phytochem.2005.08.001
Koo HJ, Gang DR (2012) Suites of terpene synthases explain differential terpenoid production in ginger and turmeric tissues. PLoS ONE 7:e51481. https://doi.org/10.1371/journal.pone.0051481
Li Y, Smolke CD (2016) Engineering biosynthesis of the anticancer alkaloid noscapine in yeast. Nat Commun 7:12137. https://doi.org/10.1038/ncomms12137
Li S, Yuan W, Deng G, Wang P, Yang P, Aggarwal BB (2011) Chemical composition and product quality control of turmeric (Curcuma longa L). Pharm Crops 2:28–54. https://doi.org/10.2174/2210290601102010028
Ma X, Gang DR (2006) Metabolic profiling of in vitro micropropagated and conventionally greenhouse grown ginger (Zingiber officinale). Phytochemistry 67:2239–2255. https://doi.org/10.1016/j.phytochem.2006.07.012
Maffei ME, Gertsch J, Appendino G (2011) Plant volatiles: production, function and pharmacology. Nat Prod Rep 28:1359–1380. https://doi.org/10.1039/c1np00021g
Mathai CK (1978) The pattern of rhizome yield and their accumulation of commercially important chemical constituents in turmeric (Curcuma species), during growth and development. Qual Plant 28:219–225. https://doi.org/10.1007/BF01093951
Okamoto M, Kuwahara A, Seo M, Kushiro T, Asami T, Hirai N, Kamiya Y, Koshiba T, Nambara E (2006) CYP707A1 and CYP707A2, which encode abscisic acid 8′-hydroxylases, are indispensable for proper control of seed dormancy and germination in arabidopsis. Plant Physiol 141:97–107. https://doi.org/10.1104/pp.106.079475
Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main A, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Dorin G, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532. https://doi.org/10.1038/nature12051
Rai A, Saito K, Yamazaki M (2017) Integrated omics analysis of specialized metabolism in medicinal plants. Plant J 90:764–787. https://doi.org/10.1111/tpj.13485
Rather GA, Sharma A, Pandith SA, Kaul V, Nandi U, Misra P, Lattoo SK (2018) De novo transcriptome analyses reveals putative pathway genes involved in biosynthesis and regulation of camptothecin in Nothapodytes nimmoniana (Graham) Mabb. Plant Mol Biol 96:197–215. https://doi.org/10.1007/s11103-017-0690-9
Rischer H, Oresic M, Seppanen-Laakso T, Katajamaa M, Lammertyn F, Ardiles-Diaz W, Van Montagu MC, Inze D, Oksman-Caldentey KM, Goossens A (2006) Gene-to-metabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc Natl Acad Sci USA 103:5614–5619. https://doi.org/10.1073/pnas.0601027103
Rodriguez S, Kirby J, Denby CM, Keasling JD (2014) Production and quantification of sesquiterpenes in Saccharomyces cerevisiae, including extraction, detection and quantification of terpene products and key related metabolites. Nat Protoc 9:1980–1996. https://doi.org/10.1038/nprot.2014.132
Taniguchi S, Miyoshi S, Tamaoki D, Yamada S, Tanaka K, Uji Y, Tanaka S, Akimitsu K, Gomi K (2014) Isolation of jasmonate-induced sesquiterpene synthase of rice: product of which has an antifungal activity against Magnaporthe oryzae. J Plant Physiol 171:625–632. https://doi.org/10.1016/j.jplph.2014.01.007
Wisecaver JH, Borowsky AT, Tzin V, Jander G, Kliebenstein DJ, Rokas A, Wisecaver JH (2017) A global coexpression network approach for connecting genes to specialized metabolic pathways in plants. Plant Cell 29:944–959. https://doi.org/10.1105/tpc.17.00009
Xia J, Wishart DS (2016) Using metaboanalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinform 55:14101–141091. https://doi.org/10.1002/cpbi.11
Xie Z, Ma X, Gang DR (2009) Modules of co-regulated metabolites in turmeric (Curcuma longa) rhizome suggest the existence of biosynthetic modules in plant specialized metabolism. J Exp Bot 60:87–97. https://doi.org/10.1093/jxb/ern263
Xu J, Ji F, Kang J, Wang H, Li S, Jin DQ, Zhang Q, Sun H, Guo Y (2015) Absolute configurations and NO inhibitory activities of terpenoids from Curcuma longa. J Agric Food Chem 63:5805–5812. https://doi.org/10.1021/acs.jafc.5b01584
Yadav D, Yadav SK, Khar RK, Mujeeb M, Akhtar M (2013) Turmeric (Curcuma longa L): A promising spice for phytochemical and pharmacological activities. Int J Green Pharm 7:85–89. https://doi.org/10.22377/ijgp.v7i2.302
Zhang Y, Li ZX, Yu XD, Fan J, Pickett JA, Jones HD, Zhou JJ, Birkett MA, Caulfield J, Napier JA, Zhao GY, Cheng XG, Shi Y, Bruce TJ, Xia LQ (2015) Molecular characterization of two isoforms of a farnesyl pyrophosphate synthase gene in wheat and their roles in sesquiterpene synthesis and inducible defence against aphid infestation. New Phytol 206:1101–1115. https://doi.org/10.1111/nph.13302
Zhuang X, Kollner TG, Zhao N, Li G, Jiang Y, Zhu L, Ma J, Degenhardt J, Chen F (2012) Dynamic evolution of herbivore-induced sesquiterpene biosynthesis in sorghum and related grass crops. Plant J 69:70–80. https://doi.org/10.1111/j.1365-313X.2011.04771.x
Acknowledgements
This work was supported by Grants from the National Natural Science Foundation of China (81603244, 81822046, and 81573532), China Postdoctoral Science Foundation (2015M571252), key project at central government level: The ability establishment of sustainable use for valuable Chinese medicine resources (2060302) and National Mega-Project for Innovative Drugs (2018ZX09711001-006-004). We thank Doctor Qingmiao Li and Doctor Xianjian Zhou from the Institute of Traditional Chinese Medicine Resources and Planting, Sichuan Academy of Traditional Chinese Medicine for help in collecting samples. We thank Professor Xueli Zhang and Doctor Zhubo Dai from Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences for providing the FPP production yeast strain BY-T15.
Author information
Authors and Affiliations
Contributions
LQH and JG were leading the project and designed the work. JRS, XHM, ZLZ and YM sampled the plant and did the chemical experiment. TZQ, CJSL, and WZ helped to analyze the chemical data. GHC, WG, YJZ, JFT, HZL, and YS performed gene functional characterization and bacterial engineering. YNW and TC performed transcriptome analysis. JRS, GHC, and JG wrote the paper.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, J., Cui, G., Ma, X. et al. An integrated strategy to identify genes responsible for sesquiterpene biosynthesis in turmeric. Plant Mol Biol 101, 221–234 (2019). https://doi.org/10.1007/s11103-019-00892-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00892-0