Abstract
Solanum lycopersicum and Solanum habrochaites (f. typicum) accession PI127826 emit a variety of sesquiterpenes. To identify terpene synthases involved in the production of these volatile sesquiterpenes, we used massive parallel pyrosequencing (RNA-seq) to obtain the transcriptome of the stem trichomes from these plants. This approach resulted initially in the discovery of six sesquiterpene synthase cDNAs from S. lycopersicum and five from S. habrochaites. Searches of other databases and the S. lycopersicum genome resulted in the discovery of two additional sesquiterpene synthases expressed in trichomes. The sesquiterpene synthases from S. lycopersicum and S. habrochaites have high levels of protein identity. Several of them appeared to encode for non-functional proteins. Functional recombinant proteins produced germacrenes, β-caryophyllene/α-humulene, viridiflorene and valencene from (E,E)-farnesyl diphosphate. However, the activities of these enzymes do not completely explain the differences in sesquiterpene production between the two tomato plants. RT-qPCR confirmed high levels of expression of most of the S. lycopersicum sesquiterpene synthases in stem trichomes. In addition, one sesquiterpene synthase was induced by jasmonic acid, while another appeared to be slightly repressed by the treatment. Our data provide a foundation to study the evolution of terpene synthases in cultivated and wild tomato.
Similar content being viewed by others
Introduction
Terpenoids constitute a large class of chemical compounds produced by most, if not all, living organisms. Over 23,000 different terpenoid compounds have been characterized (Sacchettini and Poulter 1997). Plants produce terpenoids that function in primary metabolism such as phytohormones (abscisic acid, gibberellins, cytokinins, and brassinosteroids), are part of photosynthetic pigments (phytol and carotenoids), electron carriers (ubiquinone) or constitute structural components of membranes (phytosterol). However, the majority of plant terpenoids are secondary, or specialized metabolites, present only in a subset of plant lineages. They can be active as direct defensive compounds, such as phytoalexins that accumulate upon pathogen infection (Akram et al. 2008). In addition, volatile terpenoids can function as indirect defensive compounds by attracting predators or parasitoids of the attacking insect (Walling 2000). The emission of different terpenoids is induced by insect herbivory (Kant et al. 2004; Olson et al. 2008; Navia-Gine et al. 2009).
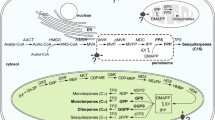
Two distinct pathways leading to the universal terpene precursors isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) operate in plants. The mevalonate (MVA) pathway produces IPP in the cytosol, which can be converted to DMAPP by IPP isomerase. The 2-C-methyl-d-erythritol 4-phosphate (MEP) pathway produces plastidial IPP and DMAPP. Head-to-tail elongation of IPP with DMAPP catalyzed by geranyl diphosphate (GPP) synthase or by neryl diphosphate (NPP) synthase lead to the formation of GPP and NPP (the (Z,Z)-isomer of GPP; Croteau and Karp 1979), respectively, which are the precursors for monoterpenes. (E,E)-Farnesyl diphosphate (FPP), and (Z,Z)-Farnesyl diphosphate (Kellog and Poulter 1997) serve as precursors for sesquiterpenes, and are synthesized by (Z,Z)- and (E,E)-FPP synthases, respectively. (E,E,E)-Geranylgeranyl diphosphate (GGPP) synthase catalyzes formation of GGPP, the diterpene precursor. The MEP pathway provides precursors for the synthesis of monoterpenes and diterpenes in plastids, whereas sesquiterpenes are derived from precursors of the MVA pathway in the cytosol. However, exchange of precursor between cytosol and plastids has been reported. Snapdragon flowers can synthesize sesquiterpenes from plastidial isoprenes, indicating transport of IPP from the plastids to the cytosol (Dudareva et al. 2005). More recently it was shown that several sesquiterpenes from wild tomato (Solanum habrochaites) are synthesized in the plastids from plastidial (Z,Z)-FPP (Sallaud et al. 2009).
The prenyl diphosphates are converted to terpenes by the action of terpene synthases (TPSs), a group of structurally and evolutionarily related enzymes (Chen et al. 2011). Induced terpenoid synthesis is often correlated with induced expression of terpene synthases (Zulak et al. 2009; Navia-Gine et al. 2009; Herde et al. 2008). Besides regulation at the level of terpene synthases, induction of precursor biosynthetic genes has also been reported (Kant et al. 2004; Ament et al. 2004). In vitro, sesquiterpene synthases can often produce monoterpenes when provided with GPP as substrate, and monoterpene synthases can produce sesquiterpenes when provided with FPP. Therefore, subcellular targeting of terpene synthases determines which substrate the terpene synthase encounters. For instance, two nearly identical terpene synthases from snapdragon both catalyze the conversion of GPP to linalool and of FPP to nerolidol in vitro. However, only one of these linalool/nerolidol synthases has a transit peptide, localizing the protein to the plastids (Nagegowda et al. 2008).
To be effective against pests and diseases, defensive terpenoid compounds are often produced at the surface of the plant. Trichomes, which are specialized secretory structures on the surface of leaves and stems, contain high levels of terpenes in many species (Chatzivasileiadis et al. 1999; Besser et al. 2009), as well as other secondary metabolites (van Schie et al. 2007; Fridman et al. 2005; Ben-Israel et al. 2009). Furthermore, many investigations have shown that these compounds, including terpenes, are usually synthesized de novo in the trichomes (Maes et al. 2011; Olsson et al. 2009).
New terpene synthases are often discovered by homology-based cloning (van Der Hoeven et al. 2000; Portnoy et al. 2008; van Schie et al. 2007; Jones et al. 2008), by random sequencing of cDNAs (van Der Hoeven et al. 2000; Wang et al. 2008) followed by bioinformatic searches of the resulting EST databases (Keeling et al. 2011) or by genome mining (Aubourg et al. 2002; Martin et al. 2010). Terpene synthases that are less abundant in a particular organ or structure, or have low sequence similarity to known terpene synthases might not be identified by these methods. Therefore, we set out to use massive parallel pyrosequencing of the tomato trichome transcriptome in order to find new sesquiterpene synthases present in tomato stem trichomes. The use of massive parallel pyrosequencing of transcripts, termed “RNA-seq” is becoming more widely used (Schilmiller et al. 2010; Wilhelm and Landry 2009). The advantage of RNA-seq above the construction of EST databases is that quantitative expression levels of transcripts are better estimated due to the larger set of data.
Here we describe the identification and characterization of seven sesquiterpene synthase cDNAs from Solanum lycopersicum and six sesquiterpene synthase cDNAs from Solanum habrochaites, a wild tomato species, by using RNA-seq on cDNA from stem trichomes. Functional expression in E. coli provided information on product specificity of the proteins encoded by these cDNAs. Determination of tissue-specific expression of the cultivated tomato sesquiterpene genes showed that the expression of most of them was highest in tissue containing trichomes. Furthermore, one sesquiterpene synthase in the cultivated tomato was induced by jasmonic acid treatment, suggesting its involvement in herbivore-induced terpenoid emission.
Materials and methods
Plant material and mRNA isolation
Tomato plants (Solanum lycopersicum cultivar Moneymaker and Solanum habrochaites accession PI127826) were obtained from Enza Zaden (Enkhuizen, The Netherlands) and grown in soil in a greenhouse with day/night temperatures of 23–18°C and a 16/8 h light/dark regime for 4 weeks. Cuttings were made, which were placed in soil and grown for another 3 weeks. Trichomes from the stem and petioles were collected at the bottom of a 50 ml tube by vortexing frozen petiole and stem segments in liquid nitrogen as described before (van Schie et al. 2007). RNA of trichomes was isolated using the Qiagen RNeasy plant Mini kit according to the manufacturer’s protocol. Messenger RNA was isolated from the pool of total RNA using the PolyAtract mRNA isolation system III from Promega (Madison, Wisconsin, USA) according to the manufacturers’ instructions. Messenger RNA was isolated from total RNA with an efficiency between 0.56 and 0.81%. Terpenes stored in trichomes were sampled by transferring approximately 25 mg trichomes to a 20 ml glass vial containing 2 ml saturated CaCl2 (5 M) buffered in 100 mM Na-Acetate at pH 4.5. Vials were capped immediately and kept at 8°C prior to sampling with a Solid Phase Micro Extraction fiber (SPME), as described below.
mRNA amplification and double stranded cDNA synthesis
The MessageAmp II aRNA amplification kit from Ambion (Austin, Texas, USA) was used to amplify trichome mRNA. An aliquot of 100 ng of mRNA was used as input for amplification. Amplification was carried out according to the manufacturer’s protocol. The yield of amplified anti-sense strand RNA was 171 μg for S. lycopersicum and 144 μg for S. habrochaites. The size ranged from ~400 to 3,000 bases.
First strand cDNA synthesis from the amplified RNA was carried out with random hexamer primers. Synthesis was performed in batches using 10 μg RNA in 20 μl reactions. Random primers at a final concentration of 62.5 nM were combined with amplified RNA and incubated at 70°C for 5 min in a thermo-cycler (Biometra, Göttingen, Germany) after which the sample was transferred to ice. Subsequently, RevertAid M-MulV H− reverse transcriptase from Fermentas Life Sciences (St. Leon-Rot, Germany) in combination with the supplied buffer was added with 1 mM nucleotides. First strand cDNA synthesis was carried out at 42°C for 90 min. Directly thereafter, reactions were transferred to ice and second strand synthesis was performed using Fermentas RNase H and DNA polymerase I from E. coli. To each 20 μl reaction tube 8 μl of the supplied DNA polymerase I reaction buffer was added along with 1 unit RNase H and 30 units DNA polymerase I. The reaction volume was increased to 100 μl with water. All components were added cold and the second strand synthesis reaction was carried out at 15°C for 2 h. Double stranded cDNA was purified with the QIAquick PCR purification kit (Qiagen, Germany) and subsequently used as input for massive parallel pyrosequencing.
Massive parallel pyrosequencing and data analysis
The cDNA samples were analyzed by massive parallel pyrosequencing using a Genome Sequencer GS20 sequencing platform (Roche Applied Science). First, 6 μg of each sample was subjected to nebulisation. Further library preparation was performed according to the standard GS20 library preparation protocol as supplied by Roche Applied Bioscience. Emulsion PCR and bead enrichment were carried out according to the standard GS20 protocol. One full picotiterplate (PTP; 70 × 75 mm) with two regions was used. Enriched beads were divided over both regions. Sequencing was performed according to the manufacturer’s instructions (Roche Applied Science). GS20 data processing was performed on-rig using the standard GS software, resulting in an average read length of 89.7 nt, a total number of raw reads of 768,329 and a total number of Passed Filter reads 377,673.
The Passed Filter reads (377,673 ESTs) were cleaned from low quality regions using SeqClean (http://www.tigr.org/tdb/tgi/software/). Four sequences shorter than 40 nucleotides and 422 sequences that consisted mostly of low-complexity successions were discarded. The resulting 377,447 high quality sequences were analyzed using the TGI Clustering (TGICL), a software system for fast clustering of large EST datasets (http://www.tigr.org/tdb/tgi/software/). The parameters used for clustering allowed to group sequences sharing a minimum of 95% identity over at least 40 nucleotides with less than 10 bases of mismatched sequence at either end (Quackenbush et al. 2000). Initially the reads were searched for redundant sequences and after the megablast of the TGICL, one cluster with in total 77,050 ESTs was discharged. The reads of this cluster showed high similarity to chloroplast sequences. Computational limitations did not allow for contig building of a cluster with this redundancy. The resulting sequences were searched for repeats using the dicots repeat databases available at http://www.tigr.org/tdb/e2k1/plant.repeats/. However, the trichome-derived ESTs did not show significant similarity to these repeats, and the repeat masking step of the unigene creation was omitted. A second cycle of unigene creation was performed using the output of the first cycle as input. This process created a set of merged contigs constructed by merging less than 5% of the unigenes from the first run of TGICL. A third cycle of unigene creation did not result in additional merges. The output of the second cycle contained 18,918 unigenes with 2 or more ESTs per unigene and 42,601 singlets.
cDNA library construction and isolation of full length cDNAs of terpene synthases
cDNA libraries were constructed using the HybriZAP −2.1 XR library construction kit and HybriZAP −2.1 XR cDNA synthesis kit from Stratagene (Cedar Creek, Texas, USA). The size of the primary cDNA library was 3.0 × 106 pfu for S. lycopersicum and 9.9 × 106 for S. habrochaites. The primary libraries were amplified according to the manufacturer’s protocol. The amplified cDNA libraries were excised using the Mass Excision Protocol described by the manufacturer. Excised cDNA libraries were used to PCR amplify 3′ and 5′ fragments of cDNAs of interest identified previously in the pyrosequencing database. When necessary, re-amplification was performed using nested primers. After sequence verification, the resulting full length candidate-sesquiterpene synthases were cloned in-frame into the pGEX-KG expression vector (Guan and Dixon 1991). In addition to the cDNAs from S. habrochaites accession PI127826 as described above, we obtained cDNAs from a different S. habrochaites accession (PI126449) using a published EST database (Fridman et al. 2005). Sequence data can be found in the GenBank database (http://www.ncbi.nlm.nih.gov/) under accession numbers: ShTPS9, JN402388; ShTPS12, JN402389; ShTPS14a, JN402390; ShTPS17, JN402391; ShTPS14b, JN402392; ShTPS15b, JN402393; SlTPS16, JN402394; SlTPS17, JN402395; SlTPS31, JN402396; ShTPS15a, JN402397; ShTPS16, JN402398; SlTPS15, JN402399.
Functional expression analysis
Expression constructs were transformed to C41 (DE3) electro competent E. coli cells (Dumon-Seignovert et al. 2004). From a single colony an overnight culture was grown at 37°C of which 500 μl was inoculated in 50 ml Terrific Broth containing 100 μg ml−1 ampicillin. The culture was grown to an OD600 of 0.5–0.6 at 37°C after which they were placed at 4°C for 30 min. Protein expression was induced with 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG; Roche, Basel, Switzerland). An empty plasmid was taken along as a negative control. After incubation for 16 h at 16°C at 200 rpm, cells were harvested by centrifugation at 4,500 rpm at 4°C for 15 min. The supernatant was removed and the pellet was resuspended in 2 ml of assay buffer (25 mM HEPES, pH 7.2, 10 mM MgCl2, 10% (v/v) glycerol) with added lysozyme (1 mg ml−1) and protease inhibitors (Complete, EDTA-free; Roche, Basel Switzerland). Cells were incubated on ice for 30 min after which they were sonicated. Lysates were centrifuged at 4°C for 25 min at 12,000 g. The supernatant was aliquoted and stored at −80°C.
Activity assays were performed in 20 ml glass vials in a total volume of 500 μl containing 50 mM HEPES, pH7.2, 100 mM KCl, 7.5 mM MgCl2, 20 μM MgCl2, 5% (v/v) glycerol, 5 mM DTT with 50 μl protein extract and either 2 mM (E,E)-FPP (E–E-farnesyl diphosphate), (Z,Z)-FPP (2Z-6Z-farnesyl diphosphate) or GPP (geranyl diphosphate) as substrate (Echelon Biosciences Incorporated, Salt Lake City, USA). Vials were immediately closed with a Teflon lined crimp cap and incubated under moderate shaking for 1 h at 30°C. Enzyme products were sampled with a Solid Phase Micro Extraction fiber (SPME) for 10 min after the vial had been agitated and heated to 50°C. The fiber was desorbed for 1 min in an Optic injector port (ATAS GL Int. Zoeterwoude, NL) which was kept at 250°C. Compounds were separated on a DB-5 column (10 m × 180 μm, 0.18 μm film thickness; Hewlett Packard) in an 6890 N gas chromatograph (Agilent, Amstelveen, NL) with a temperature program set to 40°C for 1.5 min, ramp to 250°C at 30°C per minute and 250°C for an additional 2.5 min. Helium was used as a carrier gas, the column flow set to 3 ml per minute for 2 min, and to 1.5 ml per minute thereafter. Mass spectra were generated with the ion source set to −70 V at 200°C and collected with a Time-of-Flight MS (Leco, Pegasus III, St. Joseph, MI, USA) at 1,850 V, with an acquisition rate of 20 scans per second. Because some terpenes, such as germacrenes, are prone to thermal conversion (Colby et al. 1998; Faraldos et al. 2007) the enzyme assays were also extracted with pentane which was injected at 50°C in the injection port. For this, 500 μl lysate was assayed in the presence of 5 mM DTT and either (E,E)-FPP, (Z,Z)-FPP or GPP as a substrate, overlaid with 2 ml pentane (Sigma). After 1 h of incubation at 30°C the pentane layer was transferred to a 2 ml glass vial and concentrated on ice under nitrogen gas, to a final volume of 50 μl. Terpenoids were analysed by injection of 2 μl into the Optic injection port (ATAS GL international) at 50°C, subsequently heated to 275°C at a rate of 4°C s−1 followed by gas-chromatography and mass-spectrometry as described by Bleeker et al. (2009). Boiled protein extract could not convert any of the precursors and was taken along as a negative control. No products were formed with GGPP as substrate. Terpene products were identified using standards when possible or by comparing mass spectra, and Kovats Index (Adams 2002).
The enzymatic assays for ShTPS15 from PI126449 and SlTPS31 from cultivar M82 and the analyses of their products were performed as described in Falara et al. (in press).
Expression analysis
RNA was isolated using TRIzol (Invitrogen, Paisley, UK) according to the manufacturer’s instructions, from different tissues of 4-week-old S. lycopersicum plants. RNA was isolated from leaves, whole stem (stem pieces with trichomes), bald stem (stem pieces with trichomes removed), stem trichomes, root, fruit and flowers. Additionally, RNA was isolated from stem trichomes of 4-week-old S. lycopersicum plants 24 h after spraying with either 1 mM jasmonic acid (Ducheva Biochemicals) in tap water containing 0.05% SilwetL-77, or water and SilwetL-77 alone.
The RNA was DNase treated (TURBO DNase kit, Ambion) and the quantity determined with a Nanodrop spectrofotometer (ND-1000, Thermo Scientific). First strand cDNA was synthesized from 1.5 μg of total RNA using the RevertAid kit from Fermentas according to the manufacturers’ instructions. For expression analysis the cDNA equivalent of 100 ng RNA was used as template with the SYBR Green PCR qPCR SuperMix UDG (Invitrogen) and 300 nM of each primer and dispersed as 20 μl on a 96-well optical reaction plate (Applied Biosystems). PCRs were performed in the ABI 7500 Real-Time PCR System (Applied Biosystems). The specificity of the reaction was verified by dissociation analysis. Expression of ACTIN (SGN-U579547) was used to normalize and correct for variance in quality of RNA and quantity of input cDNA. Primer pair efficiencies were estimated by analysis of amplification curves of a standard cDNA dilution range. Three biological replicates were analyzed individually and statistical significance was tested by ANOVA.
Results
Sequences of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites show high levels of identity
Massive parallel pyrosequencing of the trichome transcriptomes resulted in 195,377 reads from cDNA derived from S. lycopersicum and 182,386 reads from S. habrochaites accession PI127826, both with an average length of 80 nucleotides. About 0.20% of all reads had high sequence similarity to known sesquiterpene synthases for S. lycopersicum versus 0.12% for S. habrochaites reads (Table 1). This resulted in the identification of multiple TPS sequences belonging to the TPS-a clade according to the phylogeny of the plant TPS family (Bohlmann et al. 1998; Chen et al. 2011). Recently we have analyzed the nearly completed genomic sequence of S. lycopersicum, and have identified a total of 44 TPS genes (Falara et al. in press). This analysis also showed that the majority of TPS genes in the tomato genome appear to encode sesquiterpene synthases and most of these putative sesquiterpene synthases belong to the TPS-a clade. In the S. lycopersicum database, transcripts for TPS9 (previously shown to encode germacrene C synthase; van Der Hoeven et al. 2000), TPS12 (previously shown to encode β-caryophyllene/α-humulene synthase; van Der Hoeven et al. 2000), TPS15, TPS16 and TPS17 were identified, as well as low numbers of reads for TPS31 (TPS31 was formerly known as LeVS2 (GenBank: AAG09949.1), but the function of LeVS2 was not established). It was noted that the full-length cDNA of TPS15 has a premature stop at position 228 and is predicted to encode a non-functional protein of 76 amino acids.
Multiple putative sesquiterpene synthases from S. habrochaites accession PI127826 were also observed, and full-length cDNAs for these genes were isolated. Based on phylogenetic analysis of their DNA sequences (Fig. 1), these cDNAs came from the TPS9, TPS12, TPS15, and TPS17. Again, TPS15 had several premature stop codons in the 5′ region of the cDNA but could putatively encode a shorter protein of 499 amino acids if the ATG at position 157 is functional as start codon, albeit this protein is unlikely to have sesquiterpene synthase activity. We were able to use the primers for S. lycopersicum TPS16 to amplify the S. habrochaites TPS16 cDNA as well. However, the ShTPS16 cDNA contained a single nucleotide deletion at position 459, a mutation that results in a premature stop at amino acid 153, thus ShTPS16 is predicted to encode a non-functional enzyme.
Phylogenetic tree of terpene synthase cDNAs from the cultivated tomato S. lycopersicum (cv. Moneymaker) and the wild tomato S. habrochaites (PI127826). Open reading frames from the cDNAs were aligned with CLUSTALW. UTR sequences were not included. The phylogenetic tree was constructed after bootstrap analysis (n = 1,000) with a cut-off value of 60% using Lasergene DNAstar Megalign software (DNASTAR, Madison, USA). Copalyl diphosphate synthase (SlTPS40), involved in gibberellic acid biosynthesis forms the out-group (Chen et al. 2011)
Searching the trichome EST database of S. habrochaites accession PI126449, a line whose trichomes contain high levels of methylketones but also some sesquiterpenes (Fridman et al. 2005), identified a sequence in the TPS-a clade with high similarity to SlTPS15 (Suppl. Fig. 1) and additionally a sequence with high similarity to SlTPS14 (Falara et al. in press). The full-length cDNA of the ShTPS15 had an open reading frame of 540 codons without any apparent mutations. Apart from the sequence of TPS14 from S. lycopersicum, expressed primarily in roots (Fig. 7), we were able to isolate a full-length cDNA of ShTPS14 from stem trichomes of accession PI127826 and flowers of accession PI126449. These two ShTPS14 cDNAs had an open reading frame putatively encoding proteins of 554 amino acids.
Phylogenetic analysis (Fig. 1) of the newly identified and previously characterized tomato TPSs shows that SlTPS9 clusters together with previously identified germacrene synthases from cultivated tomatoes (SlVFNT, SSTLE1, SSTLE2) and ShTPS9 with germacrene synthases from S. habrochaites accession LA1777 (SSTLH1 and SSTLH2). It also indicates that each TPS from S. lycopersicum has a higher similarity to its respective ortholog in S. habrochaites accession PI127826 than to other tomato TPSs. Both monoterpene synthases TPS5 and TPS4 (formerly MTS1 and 2; van Schie et al. 2007) cluster together in the phylogenetic tree, as well as santalene/bergamotene (sesquiterpene) synthase (ShSBS) from S. habrochaites, which accepts (Z,Z)-FPP as a substrate (Sallaud et al. 2009), and phellandrene (monoterpene) synthase (TPS20, formerly SlPHS1) from cultivated tomato, which mainly uses NPP as substrate (Schilmiller et al. 2009). The relationships between the different TPSs were confirmed by a phylogenetic tree based on the amino acid sequences (Suppl. Fig. 2). Alignment of the deduced amino acid sequences of TPS9, TPS12, TPS16 and TPS17, depicted in Supplemental figure 3, shows how closely related TPS9 and TPS12, and TPS16 and TPS17, respectively, are. All proteins contained the RR/P8xW motive found in most terpene synthases, which is involved in hydrolysis of the pyrophosphate group and the DDxxD motive, involved in cofactor binding.
Enzymatic activity of recombinant sesquiterpene synthases
Proteins isolated from E. coli cells expressing the putative sesquiterpene synthases were assayed for their ability to convert (E,E)-FPP into sesquiterpenes. The protein encoded by the Moneymaker allele of SlTPS9 produced germacrene C, and minor amounts of germacrene A, B and D (Fig. 2a), similar to the protein encoded by this locus in cultivar VFNT Cherry tomato. ShTPS9 catalyzed the formation of mostly germacrene B and minor amounts of A and C (Fig. 2b), similar to the products observed in the reaction catalyzed by a very similar protein, designated SSTLH1 encoded by a cDNA isolated from S. habrochaites accession LA1777 (van Der Hoeven et al. 2000). Both recombinant TPS12 proteins had β-caryophyllene/α-humulene synthase activity (Fig. 3a, b).
Enzymatic activity of recombinant TPS9. GC–MS chromatogram of a S. lycopersicum (Moneymaker) and b S. habrochaites (PI127826) sesquiterpene products produced by TPS9 ectopically expressed in E. coli, assayed with (E,E)-FPP. Terpenes were extracted in pentane. Sesquiterpene peaks 1 germacrene D, 2 germacrene A, 3 germacrene C, 4 germacrene B. The chromatogram shows the detector response for the terpene-specific ion mass 93
Enzymatic activity of recombinant TPS12 and 17. GC–MS chromatograms of S. lycopersicum (Moneymaker) and S. habrochaites (PI127826) sesquiterpene products produced by TPS12 or 17 ectopically expressed in E. coli, assayed with (E,E)-FPP and measured by Solid Phase Microextraction (SPME) sampling. a SlTPS12 b ShTPS12 c SlTPS17 and d ShTPS17. Sesquiterpenes peaks: 1 β-caryophyllene, 2 α-humulene, 3 (E)-β-farnesene, 4 γ-gurjunene, 5 valencene, 6 (E,E)-α-farnesene, asterisk unidentified. The chromatogram shows the detector response for the terpene-specific ion mass 93
SlTPS17 and ShTPS17, which are 98.6% identical and 99.1% similar on the protein level, produced both mostly valencene from (E,E)-FPP and also an unidentified sesquiterpene, besides azulene and α- and β-farnesenes (SlTPS17; Fig. 3c) or β-farnesene (ShTPS17; Fig. 3d). As predicted, SlTPS15 and ShTPS15 from accession PI127826 had no activity, but the ShTPS15 allele isolated from accession PI126449 encoded a protein that catalyzed the formation of mostly germacrene A (Fig. 4). We were unable to detect activity of SlTPS16 although a soluble protein of the correct size was expressed in E. coli upon IPTG induction (data not shown). Remarkably, all recombinant proteins that could use (E,E)-FPP as substrate could also use (Z,Z)-FPP as substrate (Suppl. Fig. 4). SlTPS9 made mostly germacrene C from both (E,E)-FPP as well as (Z,Z)-FPP whereas ShTPS9 produced germacrene B and α-humulene with (Z,Z)-FPP (Suppl. Fig. 4a,b). SlTPS12 and ShTPS12 both made curcumene and β-bisabolene from (Z,Z)-FPP (Suppl. Fig. 4c,d). SlTPS17 and ShTPS17 made various bisabolenes when activity assays were done with (Z,Z)-FPP (Suppl. Fig. 4e,f).
Enzymatic activity of recombinant TPS15. GC–MS chromatograms of S. habrochaites (PI126449) sesquiterpene product produced by TPS15 ectopically expressed in E. coli assayed with (E,E)-FPP and measured by Solid Phase Microextraction (SPME) sampling. The main peak is β-elemene, the heat degradation product of germacrene A. The chromatogram shows the detector response for total ion current
Although the TPS14 proteins only differ in a few amino acids from each other (Suppl. Fig. 5), their capacity to use (Z,Z)- or (E,E)-FPP as substrate differed substantially. SlTPS14 could use either (E,E)-FPP or (Z,Z)-FPP equally well to make mostly (Z)-γ-bisabolene or α-bisabolene, respectively (Falara et al. in press). Interestingly, ShTPS14 from PI126449 appears to have no clear preference for either (Z,Z)-FPP or (E,E)-FPP. In the presence of (Z,Z)-FPP the enzyme made predominantly (Z)-β-farnesene, α- and β-acoradiene and α- and γ-bisabolenes, while when assayed with (E,E)-FPP α-cederene, (Z)-thujopsene, β-farnesene, β-acoradiene and β-bisabolene were produced (Fig. 5). ShTPS14 from PI127826 on the other hand clearly favored (E,E)-FPP to synthesize β-farnesene and α- and β-bisabolene (Fig. 5a, b). SlTPS31 predominantly made viridiflorene from (E,E)-FPP (Fig. 6).
Enzymatic activity of recombinant TPS14. GC–MS chromatograms of sesquiterpene products produced by S. habrochaites TPS14 from a PI127826 and b PI126449 ectopically expressed in E. coli, assayed with either (E,E)-FPP or (Z,Z)-FPP as substrate and measured by Solid Phase Microextraction (SPME). Sesquiterpene peaks: 1 α-cedrene, 2 (Z)-β-farnesene, 3 (E)-β-farnesene, 4 α-acoradiene, 5 β-acoradiene, 6 β-bisabolene, 7 (Z)-γ-bisabolene, 8 β-sesquiphellandrene, 9 (E)-γ-bisabolene, 10 (E)-α-bisabolene, 11 (Z)-thujopsene, 12 selinene, 13 (Z)-α-bisabolene, asterisk unidentified. The chromatogram shows the detector response for the terpene-specific ion mass 93
Enzymatic activity of recombinant TPS31. GC–MS chromatograms of S. lycopersicum (M82) sesquiterpene product produced by TPS31 ectopically expressed in E. coli assayed with (E,E)-FPP and measured by Solid Phase Microextraction (SPME) sampling. The sesquiterpene product is viridiflorene. The chromatogram shows the detector response for total ion current
All sesquiterpene synthases produce monoterpenes from GPP
All sesquiterpene synthases were assayed for monoterpene synthase activity with GPP as substrate. Though not at high efficiency, they all converted the GPP precursor to a range of most simple monoterpenes (Suppl. Fig. 6). TPS9 and TPS12 produced mostly β-myrcene, limonene and low amounts of terpinolene. In the assays with TPS17 and ShTPS14 of PI126449, additionally the monoterpenes (Z)-β-ocimene, (E)-β-ocimene and linalool were produced (Suppl. Fig. 6).
Expression of S. lycopersicum sesquiterpene synthases is differentially regulated
To investigate in which tissues the active S. lycopersicum sesquiterpene synthases TPS9, TPS12, TPS14, TPS17 and TPS31 were transcribed, we dissected mature S. lycopersicum plants for RNA isolation and subsequent quantitative RT-PCR. Expression of all terpene synthases was lowest in fruits (Fig. 7). TPS9 and TPS17 displayed similar expression patterns, with highest expression in stem trichomes (Fig. 7a, d). TPS12 expression was highest in leaves (Fig. 7b), most likely in the leaf trichomes as shown for SlTPS12 in the cultivar M82 (Schilmiller et al. 2010). In S. lycopersicum, TPS14 is expressed mostly in the roots (Fig. 7c), while in the S. habrochaites accession PI127826 TPS14 transcripts were found in trichomes (data not shown). Expression of TPS31 was very low overall, but highest in stem trichomes and leaves (Fig. 7e), most likely in their trichomes.
Tissue specific expression of SlTPSs. Relative transcript levels for a SlTPS9, b SlTPS12, c SlTPS14, d SlTPS17 and e SlTPS31 as determined by Q-RT-PCR. Mean values (+SE) of 3 biological replicas are shown, normalized for Actin expression. L leaf, WS whole stem, BS bald stem, T stem trichomes, R root, Fr fruit, Fl flower
Differential expression in response to jasmonate treatment
Since jasmonic acid (JA) treatment resulted in the induction of SlMTS1 (SlTPS3) in stem trichomes (van Schie et al. 2007), we tested whether expression of SlTPS9, SlTPS12, SlTPS17 or SlTPS31 was induced by JA treatment. Moneymaker plants that were 4 weeks old were treated with 1 mM JA or left untreated as a control, RNA was isolated after 24 h, and the levels of transcripts were measured by qRT-PCR (Fig. 8). Only the trichome-specific expression of SlTPS31 was significantly induced, approximately threefold by JA treatment, just as the positive control SlMTS1 (SlTPS3, data not shown). Interestingly, expression of SlTPS17 appeared to be reduced roughly twofold in JA-treated plants (P = 0.059).
JA induction of SlTPSs expressed in the stem trichomes. Relative transcript levels for a SlTPS9, b SlTPS12, c SlTPS17 and d SlTPS31 as determined by Q-RT-PCR. Expression in isolated stem trichomes of control (C) and JA-treated plants (JA) are shown as mean values (+SE) of 3 biological replicas, normalized for Actin expression. Asterisks indicate significant difference (P < 0.05)
Discussion
Large scale transcript sequencing as tool for gene discovery
The presence and biosynthesis of sesquiterpenes in S. lycopersicum and S. habrochaites had been previously investigated (Colby et al. 1998; Schilmiller et al. 2010; van Der Hoeven et al. 2000). Colby et al. (1998) isolated a cDNA from the cultivar VNFT Cherry and showed that it encodes germacrene C and van der Hoeven et al. (2000) showed that synthesis of germacrene C, β-caryophyllene and α-humulene was controlled by a locus on chromosome 6. Using the VFNT cDNA of germacrene C as a probe to screen cDNA libraries, van der Hoeven et al. (2000) also isolated two cDNAs, designated SSTLE1 and SSTLE2, that were very similar to VNFT germacrene C. Based on the sequences of both cDNAs, we can conclude that they are different alleles (or one of them may be a cloning artifact) of SlTPS9, which we now know to be located on chromosome 6 (Falara et al. in press). Van der Hoeven et al. (2000) also identified two alleles of TPS9 from S. habroachaites accession LA1777, designated SSTLH1 and SSTLH2, whose cDNAs encode germacrene B and germacrene D, respectively.
Subsequently, Schilmiller et al. (2010), using a proteomic approach, identified a protein sequence in S. lycopersicum cv. M82 trichomes that catalyzed the formation of β-caryophyllene and α-humulene, and was therefore designated as CAHS (β-caryophyllene/α-humulene synthase). Our data indicate that TPS12 encodes CASH. Here we used massive parallel pyrosequencing as an approach for terpene synthase transcript discovery in a specialized tissue type enriched for plant secondary metabolites, the trichomes of wild and cultivated tomato plants. Using this approach, we were able to identify transcripts of not only TPS9 and TPS12 but also of TPS14, TPS15, TPS16, TPS17 and TPS31 in the trichomes of S. lycopersicum, S. habrochaites, or both.
Over representation of germacrene synthases
Germacrene synthases have been isolated from species such as poplar (Arimura et al. 2004), goldenrod (Prosser et al. 2004), kiwi fruit (Nieuwenhuizen et al. 2009), melon (Portnoy et al. 2008), Cistus creticus (Falara et al. 2008) and many others. Often when a gene discovery approach is used to find new terpene synthases, in vitro functional assays show that multiple genes in a species encode germacrene synthases. For example, a degenerate primer strategy to find sesquiterpenes in lettuce resulted in the identification of only two germacrene A synthases (Bennett et al. 2002). Recently five sesquiterpenes from the fungus Coprinus cinereus were isolated and two of these recombinant sesquiterpene synthases catalyze the formation of germacrene A (Agger et al. 2009). Similarly, two out of three characterized sesquiterpene synthases from sunflower were identified as germacrene A synthases (Gopfert et al. 2009). The observation that three of the sesquiterpene synthases from tomato that we and others have characterized—SlTPS9 and ShTPS9, and ShTPS15 from accession PI126449—produce germacrenes with FPP as the in vitro substrate is remarkable, since only minor amounts of volatile germacrenes are present in trichomes (Kang et al. 2010; Paetzold et al. 2010; van Der Hoeven et al. 2000) or emitted from tomato (Ament et al. 2004; Kant et al. 2004). It has been postulated that germacrene A is an intermediate in epi-aristolochene synthase catalysed by tobacco 5-epi-aristolochene synthase (Rising et al. 2000). Moreover, certain sesquiterpenes in Medicago trunculata were proven to be generated predominantly via protonation of the neutral intermediate, germacrene D, as opposed to synthesis directly via FPP isomerization to nerolidyl diphosphate (Garms et al. 2010). Likewise, Selina-3,7(11)-diene (Table 2) is the result of a C6 protonation of germacrene B (Davis and Croteau 2000). Germacrene A can also be derivatized by other enzymes as is suggested for germacrene A from lettuce (Bennett et al. 2002). Therefore, in planta assays are necessary to determine whether these enzymes are bona fide germacrene synthases.
All sesquiterpene synthases accepted both (E,E)-FPP and (Z,Z)-FPP as substrates
TPS14 synthases from both S. habrochaites accessions catalyzed the formation of multiple terpenes from (E,E)-FPP and (Z,Z)-FPP, including farnesenes, acoradienes and a variety of bisabolenes (Fig. 5). However, it appears that TPS14 from accession PI127826 exhibited a preference for the (cytosolic) (E,E) isoform of FPP, whereas intriguingly, the TPS14 from accession PI126449 appeared to have an equally high affinity for (Z,Z)-FPP instead. When each ShTPS14 was assayed with both FPP-isomers present, compounds derived from both isomers were identified for ShTPS14 PI126449 whereas the ShTPS14 PI127826 products were identical to the ones seen when the enzyme was incubated with (E,E)-FPP alone (data not shown). In S. lycopersicum, TPS14 accepted (E,E)-FPP and (Z,Z)-FPP equally well, producing mostly β-bisabolene and α-bisabolene, respectively (Falara et al. in press).
ShTPS14 makes multiple products, ((E)-α-bisabolene, γ-bisabolenes and β-bisabolenes and β-farnesenes; Fig. 5) which, most likely, are direct products of deprotonation of the bisabolyl cation (C6-ring) formed from FPP via nerolidol diphosphate, without other intermediates (Davis and Croteau 2000). Interestingly, the TPS14 proteins appear to contain two aspartate-rich DDxxD Mg2+-binding motifs (305 and 527; Suppl. Fig. 5) as well as a ‘protonation-initiated’ mechanism catalytic DxDD motif (in ShTPS14)(104; Suppl. Fig. 5), both of which are common in di-and tri-terpene cyclases. The presence of a second DDxxD motif could be responsible for alternate orientations of the substrate and therefore synthesis of multiple products (Davis and Croteau 2000). TPS9, 12 and 17 also accepted (Z,Z)-FPP as substrate (Suppl. Fig. 4). Our observations of sesquiterpene synthases reacting with substrates other than the canonical (E,E)-FPP adds additional weight to previous intimations of the flexibility of these enzymes. For example, Jones et al. (2011) reported a cytosolic sequiterpene synthase from sandalwood able to use both (E,E) and (Z,Z) isomers of FPP to produce similar compounds. Such observations from multiple species suggest that these properties of the enzyme are not in vitro artifacts but might have in vivo relevance. However, whether (Z,Z)-FPP is available in the cytosol of the Solanum trichomes is not yet known, although evidence has been presented that S. habrochaites accession LA1777 produces (Z,Z)-FPP in the plastids (Sallaud et al. 2009).
Albeit with low efficiency, all TPSs described here were able to convert GPP to (mono) terpene products (Suppl. Fig. 6) indicating a level of plasticity of the active pocket of the protein. While some sesquiterpene synthases that are restricted to the use of a single substrate even with regard to precursors of the same size ((E,E)-FPP or (Z,Z)-FPP; Besser et al. 2009), there are examples of terpene synthases that can accommodate both GPP as well as (E,E)-FPP in a productive manner (van Schie et al. 2007) or even GGPP as a third possible substrate (Martin et al. 2010; Arimura et al. 2008). It has been proposed that trace amounts of GPP are present in the cytosol, and minor amounts of (E,E)-FPP are available to plastid-localised terpene synthases (Aharoni et al. 2006; Wu et al. 2006). Hence, the active site plasticity of some TPS to accommodate isoprenyl diphosphates of different chain length may be biologically relevant.
Correlations between TPS transcript abundance, TPS products and sesquiterpene production in trichomes
Despite the fact that monoterpenes appear to dominate S. lycopersicum volatile emissions (Buttery et al. 1987; Bleeker et al. 2009), the majority of TPS genes mined from the tomato genome are sesquiterpene synthases (Falara et al. in press). Based on the in vitro functional characterization of the group of sesquiterpene synthases whose transcripts we detected in stem trichomes in S. lycopersicum and S. habrochaites, the presence of most, but not all of the sesquiterpenes observed in the respective glands can be explained (Table 2). Most notably, we did not find any terpene synthase that could produce zingiberene, the most abundant sesquiterpene in S. habrochaites PI127826 (Bleeker et al. 2009). Also, trichomes of S. lycopersicum contain, albeit at minor amounts, the sesquiterpenes α-copaene and cuparene that are unaccounted for in the SlTPS enzymes assayed here.
Since some sesquiterpenes are made by more than one sesquiterpene synthases in the same species, it is difficult to determine the direct contribution of each of them to the observed mixture even when the expression levels of individual sesquiterpene synthases are examined in detail. Because we used non-normalized cDNAs in the RNA-seq experiments, we were able to analyze the data output for comparison of transcript levels of the different genes between S. lycopersicum and S. habrochaites, and the results indicate that overall abundance of sesquiterpene synthase transcripts did not correlate with the total amount of emitted sesquiterpenes, as S. habrochaites PI127826 emits over 50 μg g−1 FW sesquiterpenes in 24 h (Bleeker et al. 2009) whereas S. lycopersicum plants only emits 0.7 μg sesquiterpenes g−1 FW per day and total read abundance in the RNA-seq was 240 versus 383 respectively (Table 1). Although transcript abundance need not be translated into protein abundance, another explanation for the low sesquiterpene content of S. lycopersicum might be found in low precursor biosynthesis. Quantitative RT-PCR on cDNA derived from stem trichomes of S. lycopersicum and S. habrochaites LA1777 showed that expression of HMG-CoA reductase, a key enzyme in the MEP pathway, is approx. sevenfold higher in S. habrochaites (Besser et al. 2009). Our own RNA-seq data also show that transcript levels of HMG-CoA reductase are much higher (180 reads) in S. habrochaites than in S. lycopersicum (24 reads). It is possible that the in planta function of terpene synthases may differ from the in vivo function we found in our enzymatic assays though a more likely explanation for the discrepancy between the total sesquiterpene emission and the level of expression of synthases found might be that we have not annotated all sesquiterpene sythase sequences in our dataset. In this study we have confined our analysis of sesquiterpene biosynthesis to sesquiterpene synthases that belong to the TPS-a clade and have no plastid-targeting transit sequences. In S. habrochaites accession LA1777, a member of the TPS-e/f clade designated SBS is known to be localized in the plastid and to catalyze the formation of the sesquiterpenes santalene and bergamotene synthase from (Z,Z)-FPP (Sallaud et al. 2009), and although our RNA-seq database contains sequences that are highly similar to SBS, we did not characterize them further.
In addition to divergence in the enzymatic properties of S. lycopersicum and S. habrochaites alleles of respective TPS genes described above, the two species also differ in the set of sesquiterpene synthase genes that are expressed in their trichomes. For example, TPS14 is expressed in S. habrochaites trichomes but not in the S. lycopersicum trichomes, whereas TPS31 is expressed in the latter but not in the former, although in both cases the expression of these genes was relatively low. Thus, we can conclude that the differences in the repertoire of sesquiterpenes produced in these two Solanum species is due both to evolution of enzymatic activity and regulation of gene expression. Finally, we were also able to show that the major product of SlTPS17 and ShTPS17 is valencene (Fig. 3). This compound is detected in S. habrochaites trichomes, but not in the S. lycopersicum trichomes (Table 2). The major product of SlTPS31, viridiflorene (Fig. 6), we could not detect in trichomes of either species (Table 2).
In conclusion, our RNAseq approach has allowed us to identify sesquiterpene synthases that are expressed in stem trichomes of cultivated and wild tomato species. Some of these genes are also expressed elsewhere in the plant, but others appear to be specifically expressed or highly enriched in stem trichomes. The analysis of the enzymatic activity of orthologs revealed that in some case different products are made by orthologous sesquiterpene synthases and that some orthologs differ in their substrate preference. These results provide new tools to study the evolution of terpene synthase activity at the level of protein structure.
References
Adams RP (2002) Identification of essential oil components by gas chromatography/quadrupole mass spectroscopy. Allured Publishing Corporation, Illinois
Agger S, Lopez-Gallego F, Schmidt-Dannert C (2009) Diversity of sesquiterpene synthases in the basidiomycete Coprinus cinereus. Mol Microbiol 72(5):1181–1195
Aharoni A, Jongsma M, Kim T-Y, Ri M-B, Giri A, Verstappen F, Schwab W, Bouwmeester H (2006) Metabolic engineering of terpenoid biosynthesis in plants. Phytochem Rev 5(1):49–58
Akram A, Ongena M, Duby F, Dommes J, Thonart P (2008) Systemic resistance and lipoxygenase-related defence response induced in tomato by Pseudomonas putida strain BTP1. BMC Plant Biol 8:113
Ament K, Kant MR, Sabelis MW, Haring MA, Schuurink RC (2004) Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol 135(4):2025–2037
Arimura G, Huber DPW, Bohlmann J (2004) Forest tent caterpillars (Malacosoma disstria) induce local and systemic diurnal emissions of terpenoid volatiles in hybrid poplar (Populus trichocarpa x deltoides): cDNA cloning, functional characterization, and patterns of gene expression of (-)-germacrene D synthase, PtdTPS1. Plant J 37(4):603–616
Arimura G, Garms S, Maffei M, Bossi S, Schulze B, Leitner M, Mithofer A, Boland W (2008) Herbivore-induced terpenoid emission in Medicago truncatula: concerted action of jasmonate, ethylene and calcium signaling. Planta 227(2):453–464
Aubourg S, Lecharny A, Bohlmann J (2002) Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol Gen Genomics 267(6):730–745
Ben-Israel I, Yu G, Austin MB, Bhuiyan N, Auldridge M, Nguyen T, Schauvinhold I, Noel JP, Pichersky E, Fridman E (2009) Multiple biochemical and morphological factors underlie the production of methylketones in tomato trichomes. Plant Physiol 151(4):1952–1964
Bennett MH, Mansfield JW, Lewis MJ, Beale MH (2002) Cloning and expression of sesquiterpene synthase genes from lettuce (Lactuca sativa L.). Phytochemistry 60(3):255–261
Besser K, Harper A, Welsby N, Schauvinhold I, Slocombe S, Li Y, Dixon RA, Broun P (2009) Divergent regulation of terpenoid metabolism in the trichomes of wild and cultivated tomato species. Plant Physiol 149(1):499–514
Bleeker PM, Diergaarde PJ, Ament K, Guerra J, Weidner M, Schutz S, de Both MT, Haring MA, Schuurink RC (2009) The role of specific tomato volatiles in tomato-whitefly interaction. Plant Physiol 151(2):925–935
Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95(8):4126–4133
Buttery RG, Ling LC, Light DM (1987) Tomato leaf volatile aroma components. J Agric Food Chem 35:1039–1042
Chatzivasileiadis EA, Boon JJ, Sabelis MW (1999) Accumulation and turnover of 2-tridecanone in Tetranychus urticae and its consequences for resistance of wild and cultivated tomatoes. Exp Appl Acarol 23(12):1011–1021
Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66(1):212–229
Colby SM, Crock J, Dowdle-Rizzo B, Lemaux PG, Croteau R (1998) Germacrene C synthase from Lycopersicon esculentum cv. VFNT cherry tomato: cDNA isolation, characterization, and bacterial expression of the multiple product sesquiterpene cyclase. Proc Natl Acad Sci USA 95(5):2216–2221
Croteau R, Karp F (1979) Biosynthesis of monoterpenes: preliminary characterization of bornyl pyrophosphate synthetase from Sage (Salvia Officinalis) and demonstration that geranyl pyrophosphate is the preferred substrate for cyclization. Arch Biochem Biophys 198(2):512–522
Davis EM, Croteau R (2000) Cyclization enzymes in the biosynthesis of monoterpenes, sesquiterpenes, and diterpenes. Topics in Current Chemistry: Biosynthesis: Aromatic Polyketides, Isoprenoids, Alkaloids, vol 209. Springer-Verlag Berlin, Germany, pp 53–95
Dudareva N, Andersson S, Orlova I, Gatto N, Reichelt M, Rhodes D, Boland W, Gershenzon J (2005) The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc Natl Acad Sci USA 102(3):933–938
Dumon-Seignovert L, Cariot G, Vuillard L (2004) The toxicity of recombinant proteins in Escherichia coli: a comparison of overexpression in BL21(DE3), C41(DE3), and C43(DE3). Protein Expr Purif 37(1):203–206
Falara V, Fotopoulos V, Margaritis T, Anastasaki T, Pateraki I, Bosabalidis AM, Kafetzopoulos D, Demetzos C, Pichersky E, Kanellis AK (2008) Transcriptome analysis approaches for the isolation of trichome-specific genes from the medicinal plant Cistus creticus subsp. creticus. Plant Mol Biol 68(6):633–651
Falara V, Akhtar T, Nguyen TTH, Spyropoulou EA, Bleeker PM, Schauvinhold I, Matsuba Y, Bonini M, Schilmiller AL, Last RL, Schuurink RC, Pichersky E (in press) The tomato (Solanum lycopersicum) terpene synthase gene family. Plant Physiol
Faraldos JA, Wu S, Chappell J, Coates RM (2007) Conformational analysis of (+)-germacrene A by variable temperature NMR and NOE spectroscopy. Tetrahedron 63(32):7733–7742
Fridman E, Wang J, Iijima Y, Froehlich JE, Gang DR, Ohlrogge J, Pichersky E (2005) Metabolic, genomic, and biochemical analyses of glandular trichomes from the wild tomato species Lycopersicon hirsutum identify a key enzyme in the biosynthesis of methylketones. Plant Cell 17(4):1252–1267
Garms S, Kollner TG, Boland W (2010) A multiproduct terpene synthase from Medicago truncatula generates cadalane sesquiterpenes via two different mechanisms. J Org Chem 75(16):5590–5600
Gopfert JC, Macnevin G, Ro DK, Spring O (2009) Identification, functional characterization and developmental regulation of sesquiterpene synthases from sunflower capitate glandular trichomes. BMC Plant Biol 9:86
Guan KL, Dixon JE (1991) Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal Biochem 192(2):262–267
Herde M, Gartner K, Kollner TG, Fode B, Boland W, Gershenzon J, Gatz C, Tholl D (2008) Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C16-homoterpene TMTT. Plant Cell 20(4):1152–1168
Jones CG, Keeling CI, Ghisalberti EL, Barbour EL, Plummer JA, Bohlmann J (2008) Isolation of cDNAs and functional characterisation of two multi-product terpene synthase enzymes from sandalwood, Santalum album L. Arch Biochem Biophys 477(1):121–130
Jones CG, Moniodis J, Zulak KG, Scaffidi A, Plummer JA, Ghisalberti EL, Barbour EL, Bohlmann J (2011) Sandalwood fragrance biosynthesis involves sesquiterpene synthases of both the terpene synthase (TPS) a and TPS-b subfamilies, including santalene synthases. J Biol Chem 286:17445–17454
Kang JH, Shi F, Jones AD, Marks MD, Howe GA (2010) Distortion of trichome morphology by the hairless mutation of tomato affects leaf surface chemistry. J Exp Bot 61(4):1053–1064
Kant MR, Ament K, Sabelis MW, Haring MA, Schuurink RC (2004) Differential timing of spider mite-induced direct and indirect defenses in tomato plants. Plant Physiol 135(1):483–495
Keeling CI, Weisshaar S, Ralph SG, Jancsik S, Hamberger B, Dullat HK, Bohlmann J (2011) Transcriptome mining, functional characterization, and phylogeny of a large terpene synthase gene family in spruce (Picea spp.). BMC Plant Biol 11:43
Kellog BA, Poulter CD (1997) Chain elongation in the isoprenoid biosynthetic pathway. Curr Opin Chem Biol 1:570–578
Maes L, Van Nieuwerburgh FC, Zhang Y, Reed DW, Pollier J, Vande Casteele SR, Inze D, Covello PS, Deforce DL, Goossens A (2011) Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol 189(1):176–189
Martin DM, Aubourg S, Schouwey MB, Daviet L, Schalk M, Toub O, Lund ST, Bohlmann J (2010) Functional annotation, genome organization and phylogeny of the grapevine (Vitis vinifera) terpene synthase gene family based on genome assembly, FLcDNA cloning, and enzyme assays. BMC Plant Biol 10:226
Nagegowda DA, Gutensohn M, Wilkerson CG, Dudareva N (2008) Two nearly identical terpene synthases catalyze the formation of nerolidol and linalool in snapdragon flowers. Plant J 55(2):224–239
Navia-Gine WG, Yuan JS, Mauromoustakos A, Murphy JB, Chen F, Korth KL (2009) Medicago truncatula (E)-beta-ocimene synthase is induced by insect herbivory with corresponding increases in emission of volatile ocimene. Plant Physiol Biochem 47(5):416–425
Nieuwenhuizen NJ, Wang MY, Matich AJ, Green SA, Chen X, Yauk YK, Beuning LL, Nagegowda DA, Dudareva N, Atkinson RG (2009) Two terpene synthases are responsible for the major sesquiterpenes emitted from the flowers of kiwifruit (Actinidia deliciosa). J Exp Bot 60(11):3203–3219
Olson DM, Davis RF, Wackers FL, Rains GC, Potter T (2008) Plant-herbivore-carnivore interactions in cotton, Gossypium hirsutum: linking belowground and aboveground. J Chem Ecol 34(10):1341–1348
Olsson ME, Olofsson LM, Lindahl AL, Lundgren A, Brodelius M, Brodelius PE (2009) Localization of enzymes of artemisinin biosynthesis to the apical cells of glandular secretory trichomes of Artemisia annua L. Phytochemistry 70(9):1123–1128
Paetzold H, Garms S, Bartram S, Wieczorek J, Uros-Gracia EM, Rodriguez-Concepcion M, Boland W, Strack D, Hause B, Walter MH (2010) The isogene 1-deoxy-D-xylulose 5-phosphate synthase 2 controls isoprenoid profiles, precursor pathway allocation, and density of tomato trichomes. Mol Plant 3(5):904–916
Portnoy V, Benyamini Y, Bar E, Harel-Beja R, Gepstein S, Giovannoni JJ, Schaffer AA, Burger J, Tadmor Y, Lewinsohn E, Katzir N (2008) The molecular and biochemical basis for varietal variation in sesquiterpene content in melon (Cucumis melo L.) rinds. Plant Mol Biol 66(6):647–661
Prosser I, Altug IG, Phillips AL, Konig WA, Bouwmeester HJ, Beale MH (2004) Enantiospecific (+)- and (-)-germacrene D synthases, cloned from goldenrod, reveal a functionally active variant of the universal isoprenoid-biosynthesis aspartate-rich motif. Arch Biochem Biophys 432(2):136–144
Quackenbush J, Liang F, Holt I, Pertea G, Upton J (2000) The TIGR gene indices: reconstruction and representation of expressed gene sequences. Nucleic Acids Res 28(1):141–145
Rising KA, Starks CM, Noel JP, Chappell J (2000) Demonstration of germacrene A as an intermediate in 5-epi-aristolochene synthase catalysis (vol 122, p 1861, 2000). J Am Chem Soc 122(27):6526–6526
Sacchettini JC, Poulter CD (1997) Creating isoprenoid diversity. Science 277(5333):1788–1789
Sallaud C, Rontein D, Onillon S, Jabes F, Duffe P, Giacalone C, Thoraval S, Escoffier C, Herbette G, Leonhardt N, Causse M, Tissier A (2009) A novel pathway for sesquiterpene biosynthesis from Z, Z-farnesyl pyrophosphate in the wild tomato Solanum habrochaites. Plant Cell 21(1):301–317
Schilmiller AL, Schauvinhold I, Larson M, Xu R, Charbonneau AL, Schmidt A, Wilkerson C, Last RL, Pichersky E (2009) Monoterpenes in the glandular trichomes of tomato are synthesized from a neryl diphosphate precursor rather than geranyl diphosphate. Proc Natl Acad Sci USA 106(26):10865–10870
Schilmiller AL, Miner DP, Larson M, McDowell E, Gang DR, Wilkerson C, Last RL (2010) Studies of a biochemical factory: tomato trichome deep EST sequencing and proteomics. Plant Physiol 153(3):1212–1223
van Der Hoeven RS, Monforte AJ, Breeden D, Tanksley SD, Steffens JC (2000) Genetic control and evolution of sesquiterpene biosynthesis in Lycopersicon esculentum and L. hirsutum. Plant Cell 12(11):2283–2294
van Schie CC, Haring MA, Schuurink RC (2007) Tomato linalool synthase is induced in trichomes by jasmonic acid. Plant Mol Biol 64(3):251–263
Walling LL (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19(2):195–216
Wang G, Tian L, Aziz N, Broun P, Dai X, He J, King A, Zhao PX, Dixon RA (2008) Terpene biosynthesis in glandular trichomes of hop. Plant Physiol 148(3):1254–1266
Wilhelm BT, Landry JR (2009) RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods 48(3):249–257
Wu SQ, Schalk M, Clark A, Miles RB, Coates R, Chappell J (2006) Redirection of cytosolic or plastidic isoprenoid precursors elevates terpene production in plants. Nat Biotechnol 24(11):1441–1447
Zulak KG, Lippert DN, Kuzyk MA, Domanski D, Chou T, Borchers CH, Bohlmann J (2009) Targeted proteomics using selected reaction monitoring reveals the induction of specific terpene synthases in a multi-level study of methyl jasmonate-treated Norway spruce (Picea abies). Plant J 60(6):1015–1030
Acknowledgments
This work was supported by KeyGene, Senter Novem, Enza Zaden, Vilmorin & Cie, Takii & Co., and De Ruiter Seeds, and the Technological Top Institute-Green Genetics grants 1C002RP (EAS) and 1CC026RP (PMB). Funding for work in the laboratory of Eran Pichersky was provided by National Science Foundation (USA) grants DBI-0604336 and IOS-1025636, and a Department of Agriculture (USA) grant 2008-35318-04541. Funding for work in the laboratory of Jörg Bohlmann was provided by the Natural Science and Engineering Research Council of Canada (Discovery Grant), Genome Canada and Genome British Columbia. Ludek Tikovsky, Harold Lemereis, and Thijs Hendrix are acknowledged for taking care of the tomato collection and Harrie Schneiders (KeyGene) for carrying out the pyrosequencing reactions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Petra M. Bleeker and Eleni A. Spyropoulou contributed equally to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2011_9813_MOESM2_ESM.tif
Supplemental figure 1. Alignment of three TPS15 homologs. SlTPS15ORF originates from S. lycopersicum (cv. Moneymaker), ShTPS15aORF originates from S. habrochaites accession PI127826, ShTPS15b originates from S. habrochaites accession PI126449. Open reading frames were aligned with CLUSTALW using the BLOSUM matrix (http://www.ch.embnet.org/software/ClustalW.html). In the SlTPS15 and ShTPS15 (PI127826) cDNAs the premature stops were replaced by an “X” and the resulting ORF was used in the alignment. Homology was visualized using Boxshade (http://www.ch.embnet.org/software/BOX_form.html) (TIFF 32972 kb)
11103_2011_9813_MOESM3_ESM.tif
Supplemental figure 2. Phylogenetic tree of terpene synthase proteins from the cultivated tomato S. lycopersicum (cv Moneymaker) and the wild species S. habrochaites (PI127826). Open reading frames were aligned with CLUSTALW using the BLOSUM matrix. The phylogenetic tree was constructed after bootstrap analysis (n = 1000) with a cut-off value of 60% using Lasergene DNAstar Megalign software. In the translated cDNAs of SlTPS15, ShTPS15 and ShTPS16 the premature stops were replaced by an “X” and the resulting ORF was used in the alignment. (TIFF 9501 kb)
11103_2011_9813_MOESM4_ESM.tif
Supplemental figure 3. Alignment of TPS9, TPS12, TPS16 and TPS17 protein sequences from the cultivated tomato S. lycopersicum (cv. Moneymaker) and the wild species S. habrochaites (PI127826). The ShTPS16ORF has been reconstructed by the insertion of an “X” for the premature stop at position 147. Conserved motifs involved in hydrolysis of the diphosphate group (RR/Px8W) and cofactor binding (DDxxD) are boxed. In black are identical amino acids, in gray similar amino acids that are conserved in >50% of the sequences. Open reading frames were aligned with CLUSTALW using the BLOSUM matrix (http://www.ch.embnet.org/software/ClustalW.html). Homology was visualized using Boxshade (http://www.ch.embnet.org/software/BOX_form.html (TIFF 45571 kb)
11103_2011_9813_MOESM5_ESM.tif
Supplemental figure 4. Enzymatic activity of recombinant TPS9, 12 and 17 assayed with (Z,Z)-FPP. GC-MS chromatograms of sesquiterpenes formed by S. lycopersicum (Moneymaker) and S. habrochaites (PI127826) terpene synthases ectopically expressed in E. coli, assayed with (Z,Z)-FPP. TPS9 products were extracted in pentane, TPS12 and TPS17 products were measured by Solid Phase Microextraction (SPME) sampling. a SlTPS9 b ShTPS9 c SlTPS12 d ShTPS12 e SlTPS17 f ShTPS17. Sesquiterpene peaks: 1 α-humulene 2 germacrene A 3 β-bisabolene 4 germacrene C 5 germacrene B 6 γ-curcumene 7 (Z)-γ-bisabolene 8 (Z)-α-bergamotene 9 (E)-γ-bisabolene 10 (E)-α-bergamotene * unidentified. The chromatogram shows the detector response for the terpene-specific ion mass 93 (TIFF 17600 kb)
11103_2011_9813_MOESM6_ESM.tif
Supplemental figure 5. Amino acid sequence alignment of TPS14 from S. lycopersicum (SlTPS14) and S. habrochaites PI127826 (ShTPS14a) and PI126449 (ShTPS14b). Conserved motifs involved in hydrolysis of the diphosphate group (RR/Px8W) and cofactor binding (DDxxD) are boxed as well as the DxDD motif. Open reading frames were aligned with CLUSTALW using the BLOSUM matrix (http://www.ch.embnet.org/software/ClustalW.html). Homology was visualized using Boxshade (http://www.ch.embnet.org/software/BOX_form.html. (TIFF 8282 kb)
11103_2011_9813_MOESM7_ESM.tif
Supplemental figure 6. Enzymatic activity of recombinant TPS9, 12, 14 and 15 assayed with GPP. GC-MS chromatograms of monoterpenes formed by S. lycopersicum (Moneymaker) and S. habrochaites (PI127826 or PI126449) terpene synthases ectopically expressed in E. coli, assayed with GPP. Products were measured by Solid Phase Microextraction (SPME) sampling. a SlTPS9 b ShTPS9 c SlTPS12 d ShTPS12 e SlTPS17 f ShTPS17 g ShTPS14a h ShTPS14b. Monoterpene peaks: 1 α-pinene 2 β-myrcene 3 delta-2-carene 4 limonene 5 α-terpinene 6 terpinolene 7 (E)-β-ocimene 8 (Z)-β-ocimene 9 γ-terpinene 10 linalool * unidentified. The chromatogram shows the detector response for the terpene-specific ion mass 93. (TIFF 20663 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Bleeker, P.M., Spyropoulou, E.A., Diergaarde, P.J. et al. RNA-seq discovery, functional characterization, and comparison of sesquiterpene synthases from Solanum lycopersicum and Solanum habrochaites trichomes. Plant Mol Biol 77, 323 (2011). https://doi.org/10.1007/s11103-011-9813-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11103-011-9813-x