Abstract
Key message
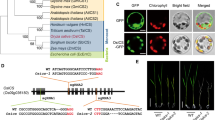
Brachypodium distachyon has a full set of exoglycosidases active on xyloglucan, including α-xylosidase, β-galactosidase, soluble and membrane-bound β-glucosidases and two α-fucosidases. However, unlike in Arabidopsis, both fucosidases are likely cytosolic.
Abstract
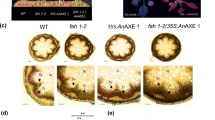
Xyloglucan is present in primary walls of all angiosperms. While in most groups it regulates cell wall extension, in Poaceae its role is still unclear. Five exoglycosidases participate in xyloglucan hydrolysis in Arabidopsis: α-xylosidase, β-galactosidase, α-fucosidase, soluble β-glucosidase and GPI-anchored β-glucosidase. Mutants in the corresponding genes show alterations in xyloglucan composition. In this work putative orthologs in the model grass Brachypodium distachyon were tested for their ability to complement Arabidopsis mutants. Xylosidase and galactosidase mutants were complemented, respectively, by BdXYL1 (Bd2g02070) and BdBGAL1 (Bd2g56607). BdBGAL1, unlike other xyloglucan β-galactosidases, is able to remove both galactoses from XLLG oligosaccharides. In addition, soluble β-glucosidase BdBGLC1 (Bd1g08550) complemented a glucosidase mutant. Closely related BdBGLC2 (Bd2g51280), which has a putative GPI-anchor sequence, was found associated with the plasma membrane and only a truncated version without GPI-anchor complemented the mutant, proving that Brachypodium also has soluble and membrane-bound xyloglucan glucosidases. Both BdXFUC1 (Bd3g25226) and BdXFUC2 (Bd1g28366) can hydrolyze fucose from xyloglucan oligosaccharides but were unable to complement a fucosidase mutant. Fluorescent protein fusions of BdXFUC1 localized to the cytosol and both proteins lack a signal peptide. Signal peptides appear to have evolved only in some eudicot lineages of this family, like the one leading to Arabidopsis. These results could be explained if cytosolic xyloglucan α-fucosidases are the ancestral state in angiosperms, with fucosylated oligosaccharides transported across the plasma membrane.






Similar content being viewed by others
References
Brennan M, Harris PJ (2011) Distribution of fucosylated xyloglucans among the walls of different cell types in monocotyledons determined by immunofluorescence microscopy. Mol Plant 4:144–156
Claverie J, Balacey S, Lemaître-Guillier C, Brulé D, Chiltz A, Granet L, Noirot E, Daire X, Darblade B, Héloir MC, Poinssot B (2018) The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front Plant Sci 9:1725
Cosgrove DJ (2018) Diffuse growth of plant cell walls. Plant Physiol 176:16–27
Crombie HJ, Chengappa S, Hellyer A, Reid JSG (1998) A xyloglucan oligosaccharide-active, transglycosylating-d-glucosidase from the cotyledons of nasturtium (Tropaeolum majus L.) seedlings—purification, properties and characterization of a cDNA clone. Plant J 15:27–38
Crombie HJ, Chengappa S, Jarman C, Sidebottom C, Reid JS (2002) Molecular characterisation of a xyloglucan oligosaccharide-acting α-d-xylosidase from nasturtium (Tropaeolum majus L.) cotyledons that resembles plant ‘apoplastic’ α-d-glucosidases. Planta 214:406–413
Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133:462–469
Dardelle F, Le Mauff F, Lehner A, Loutelier-Bourhis C, Bardor M, Rihouey C et al (2015) Pollen tube cell walls of wild and domesticated tomatoes contain arabinosylated and fucosylated xyloglucan. Ann Bot 115:55–66
de Alcantara PHN, Dietrich SMC, Buckeridge MS (1999) Xyloglucan mobilisation and purification of a (XLLG/XLXG) specific β-galactosidase from cotyledons of Copaifera langsdorffii. Plant Physiol Biochem 37:653–663
de Alcantara PHN, Martim L, Silva CO, Dietrich SMC, Buckeridge MS (2006) Purification of a β-galactosidase from cotyledons of Hymenaea courbaril L. (Leguminosae). Enzyme properties and biological function. Plant Physiol Biochem 44:619–627
Dick-Pérez M, Zhang Y, Hayes J, Salazar A, Zabotina OA, Hong M (2011) Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50:989–1000
Edwards M, Bowman YJ, Dea IC, Reid JS (1988) A β-d-galactosidase from nasturtium (Tropaeolum majus L.) cotyledons. Purification, properties, and demonstration that xyloglucan is the natural substrate. J Biol Chem 263:4333–4337
Eklof JM, Brumer H (2010) The XTH gene family: an update on enzyme structure, function, and phylogeny in xyloglucan remodeling. Plant Physiol 153:456–466
Fry SC (1994) Oligosaccharins as plant growth regulators. Biochem Soc Symp 60:5–14
Galloway AF, Pedersen MJ, Merry B, Marcus SE, Blacker J, Benning LG et al (2017) Xyloglucan is released by plants and promotes soil particle aggregation. New Phytol 217:1128–1136
Gibeaut DM, Pauly M, Bacic A, Fincher GB (2005) Changes in cell wall polysaccharides in developing barley (Hordeum vulgare) coleoptiles. Planta 221:729–738
Günl M, Pauly M (2011) AXY3 encodes a α-xylosidase that impacts the structure and accessibility of the hemicellulose xyloglucan in Arabidopsis plant cell walls. Planta 233:707–719
Günl M, Neumetzler L, Kraemer F, de Souza A, Schultink A, Pena M et al (2011) AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23:4025–4040
Hayashi T, Takeda T (1994) Compositional analysis of the oligosaccharide units of xyloglucans from suspension-cultured poplar cells. Biosci Biotechnol Biochem 58:1707–1708
Hoffman M, Jia Z, Pena MJ, Cash M, Harper A, Blackburn AR et al (2005) Structural analysis of xyloglucans in the primary cell walls of plants in the subclass Asteridae. Carbohydr Res 340:1826–1840
Hrmova M, Harvey AJ, Wang J, Shirley NJ, Jones GP, Stone B et al (1996) Barley β-d-glucan exohydrolases with β-d-glucosidase activity. Purification, characterization, and determination of primary structure from a cDNA clone. J Biol Chem 271:5277–5286
Hsieh YSY, Harris PJ (2009) Xyloglucans of monocotyledons have diverse structures. Mol Plant 2:943–965
Iglesias N, Abelenda JA, Rodino M, Sampedro J, Revilla G, Zarra I (2006) Apoplastic glycosidases active against xyloglucan oligosaccharides of Arabidopsis thaliana. Plant Cell Physiol 47:55–63
Ishimizu T, Hashimoto C, Takeda R, Fujii K, Hase S (2007) A novel α1,2-l-fucosidase acting on xyloglucan oligosaccharides is associated with endo-β-mannosidase. J Biochem 142:721–729
Kaewthai N, Gendre D, Eklöf JM, Ibatullin FM, Ezcurra I, Bhalerao R et al (2013) Group III-A XTH genes of Arabidopsis thaliana encode predominant xyloglucan endo-hydrolases that are dispensable for normal growth. Plant Physiol 161:440–454
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Lampropoulos A, Sutikovic Z, Wenzl C, Maegele I, Lohmann JU, Forner J (2013) GreenGate—a novel, versatile, and efficient cloning system for plant transgenesis. PLoS ONE 8:e83043
Lampugnani ER, Moller IE, Cassin A, Jones DF, Koh PL, Ratnayake S et al (2013) In vitro grown pollen tubes of Nicotiana alata actively synthesise a fucosylated xyloglucan. PLoS ONE 8:e77140
Léonard R, Pabst M, Bondili JS, Chambat G, Veit C, Strasser R et al (2008) Identification of an Arabidopsis gene encoding a GH95 α1,2-fucosidase active on xyloglucan oligo- and polysaccharides. Phytochemistry 69:1983–1988
Liu L, Paulitz J, Pauly M (2015) The presence of fucogalactoxyloglucan and its synthesis in rice indicates conserved functional importance in plants. Plant Physiol 168:549–560
Liu L, Hsia M, Dama M, Vogel J, Pauly M (2016) A xyloglucan backbone 6-O-acetyltransferase from Brachypodium distachyon modulates xyloglucan xylosylation. Mol Plant 9:615–617
McDougall GJ, Fry SC (1989) Anti-auxin activity of xyloglucan oligosaccharides: the role of groups other than the terminal α-l-fucose residue. J Exp Bot 40:233–238
Nakai H, Tanizawa S, Ito T, Kamiya K, Kim YM, Yamamoto T et al (2007) Function-unknown glycoside hydrolase family 31 proteins, mRNAs of which were expressed in rice ripening and germinating stages, are α-glucosidase and α-xylosidase. J Biochem 142:491–500
Park YB, Cosgrove DJ (2012) Changes in cell wall biomechanical properties in the xyloglucan-deficient xxt1/xxt2 mutant of Arabidopsis. Plant Physiol 158:465–475
Park YB, Cosgrove DJ (2015) Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol 56:180–194
Pauly M, Keegstra K (2016) Biosynthesis of the plant cell wall matrix polysaccharide xyloglucan. Annu Rev Plant Biol 67:235–259
Petersen TN, Brunak S, von Heijne G, Nielsen H (2011) SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785
Rahman M, Fujishige M, Maeda M, Kimura Y (2016) Rice α-fucosidase active against plant complex type N-glycans containing Lewis a epitope: purification and characterization. Biosci Biotechnol Biochem 80:291–294
Rui Y, Anderson CT (2016) Functional analysis of cellulose and xyloglucan in the walls of stomatal guard cells of Arabidopsis. Plant Physiol 170:1398–1419
Saffer AM (2018) Expanding roles for pectins in plant development. J Integr Plant Biol 60:910–923
Sainsbury F, Thuenemann EC, Lomonossoff GP (2009) pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnol J 7(7):682–693
Sampedro J, Pardo B, Gianzo C, Guitian E, Revilla G, Zarra I (2010) Lack of α-xylosidase activity in Arabidopsis alters xyloglucan composition and results in growth defects. Plant Physiol 154:1105–1115
Sampedro J, Gianzo C, Iglesias N, Guitián E, Revilla G, Zarra I (2012) AtBGAL10 is the main xyloglucan β-galactosidase in Arabidopsis, and its absence results in unusual xyloglucan subunits and growth defects. Plant Physiol 158:1146–1157
Sampedro J, Guttman M, Li LC, Cosgrove DJ (2015) Evolutionary divergence of β-expansin structure and function in grasses parallels emergence of distinctive primary cell wall traits. Plant J 81:108–120
Sampedro J, Valdivia ER, Fraga P, Iglesias N, Revilla G, Zarra I (2017) Soluble and membrane-bound β-glucosidases are involved in trimming the xyloglucan backbone. Plant Physiol 173:1017–1030
Scholthof KB, Irigoyen S, Catalan P, Mandadi K (2018) Brachypodium: a monocot grass model system for plant biology. Plant Cell 30:1694
Schultink A, Liu L, Zhu L, Pauly M (2014) Structural diversity and function of xyloglucan sidechain substituents. Plants 3:526–542
Sechet J, Frey A, Effroy-Cuzzi D, Berger A, Perreau F, Cueff G et al (2016) Xyloglucan metabolism differentially impacts the cell wall characteristics of the endosperm and embryo during Arabidopsis seed germination. Plant Physiol 170:1367–1380
Shigeyama T, Watanabe A, Tokuchi K, Toh S, Sakurai N, Shibuya N et al (2016) α-Xylosidase plays essential roles in xyloglucan remodelling, maintenance of cell wall integrity, and seed germination in Arabidopsis thaliana. J Exp Bot 67:5615–5629
Shinohara N, Sunagawa N, Tamura S, Yokoyama R, Ueda M, Igarashi K et al (2017) The plant cell-wall enzyme AtXTH3 catalyses covalent cross-linking between cellulose and cello-oligosaccharide. Sci Rep 7:46099
Takahashi D, Kawamura Y, Uemura M (2016) Cold acclimation is accompanied by complex responses of glycosylphosphatidylinositol (GPI)-anchored proteins in Arabidopsis. J Exp Bot 67:5203–5215
Tanthanuch W, Chantarangsee M, Maneesan J, Ketudat-Cairns J (2008) Genomic and expression analysis of glycosyl hydrolase family 35 genes from rice (Oryza sativa L.). BMC Plant Biol. 8:84
Thomson JG, Cook M, Guttman M, Smith J, Thilmony R (2011) Novel sul I binary vectors enable an inexpensive foliar selection method in Arabidopsis. BMC Res Notes 4:44
Wan JX, Zhu XF, Wang YQ, Liu LY, Zhang BC, Li GX et al (2018) Xyloglucan fucosylation modulates Arabidopsis cell wall hemicellulose aluminium binding capacity. Sci Rep 8:428
Wang T, Park YB, Cosgrove DJ, Hong M (2015) Cellulose-pectin spatial contacts are inherent to never-dried Arabidopsis primary cell walls: evidence from solid-state nuclear magnetic resonance. Plant Physiol 168:871–884
Wang T, Cheng Y, Tabuchi A, Cosgrove DJ, Hong M (2016) The target of β-expansin EXPB1 in maize cell walls from binding and solid-state NMR studies. Plant Physiol 172:2107–2119
Xiao C, Zhang T, Zheng Y, Cosgrove DJ, Anderson CT (2015) Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol 170:234–249
Zeleny R, Leonard R, Dorfner G, Dalik T, Kolarich D, Altmann F (2006) Molecular cloning and characterization of a plant α1,3/4-fucosidase based on sequence tags from almond fucosidase I. Phytochemistry 67:641–648
Ziaur Rahman M, Maeda M, Itano S, Hossain M, Ishimizu T, Kimura Y (2017) Molecular characterization of tomato α1,3/4-fucosidase, a member of glycosyl hydrolase family 29 involved in the degradation of plant complex type N-glycans. J Biochem 161:421–432
Funding
This work was supported by the Ministerio de Economía y Competividad [grant no. BIO2012-40032-C03-01]; and Xunta de Galicia [Grant Nos. GPC2013-044, ED431B 2016/021].
Author information
Authors and Affiliations
Contributions
The project was designed and conceived by JS and DR. Experiments were performed mostly by DR, with contributions by JS and ERV. DR, JS, IZ and GR analyzed the data. JS wrote the paper. All authors have revised and approved the final version of this manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rubianes, D., Valdivia, E.R., Revilla, G. et al. Xyloglucan exoglycosidases in the monocot model Brachypodium distachyon and the conservation of xyloglucan disassembly in angiosperms. Plant Mol Biol 100, 495–509 (2019). https://doi.org/10.1007/s11103-019-00875-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00875-1