Abstract
Key message
We demonstrate that the C-terminus of OsCDC48 is essential for maintaining its full ATPase activity and OsCDC48/48E interaction is required to modulate cellular processes and plant survival in rice.
Abstract
Cell division cycle 48 (CDC48) belongs to the superfamily protein of ATPases associated with diverse cellular activities (AAA). We previously isolated a rice CDC48 mutant (psd128) displaying premature senescence and death phenotype. Here, we showed that OsCDC48 (Os03g0151800) interacted with OsCDC48E (Os10g0442600), a homologue of OsCDC48, to control plant survival in rice. OsCDC48E knockout plants exhibited similar behavior to psd128 with premature senescence and plant death. Removal of the C-terminus of OsCDC48 caused altered expression of cell cycle-related genes, changed the percentage of cells in G1 and G2/M phases, and abolished the interaction between OsCDC48 itself and between OsCDC48 and OsCDC48E, respectively. Furthermore, the truncated OsCDC48–PSD128 protein lacking the C-terminal 27 amino acid residues showed a decreased level of ATPase activity. Overexpression of OsCDC48–psd128 resulted in differential expression of AAA-ATPase associated genes leading to increased total ATPase activity, accumulation of reactive oxygen species and decreased plant tiller numbers while overexpression of OsCDC48 also resulted in differential expression of AAA-ATPase associated genes leading to increased total ATPase activity, but increased plant tiller numbers and grain yield, indicating its potential utilization for yield improvement. Our results demonstrated that the C-terminal region of OsCDC48 was essential for maintaining the full ATPase activity and OsCDC48/48E complex might function in form of heteromultimers to modulate cellular processes and plant survival in rice.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The eukaryotic cell division cycle 48 (cdc48) mutant was originally isolated among a collection of cold-sensitive yeast (Saccharomyces cerevisiae) mutants defective in cell cycle (Moir et al. 1982). p97 (named after its molecular weight of 97 kDa), the mammalian homologue of CDC48, was identified in various cells/tissues of many species (Peters et al. 1990). To date, it has been shown that CDC48/p97 was involved in a wide range of cellular processes such as regulation of cell cycle progression, cell growth and proliferation, apoptosis and necrosis, ubiquitination protein degradation and membrane fusion processes in archeabacteria, yeasts, animals and plants (Golbik et al. 1999; Mouysset et al. 2008; Bègue et al. 2016; Huang et al. 2016).
CDC48/p97 belongs to the superfamily protein of ATPases associated with diverse cellular activities. A typical CDC48/p97 usually consists of an N-terminal domain, one or two copies of AAA domain, and a C-terminal domain (Dalal and Hanson 2001; Wang et al. 2004). Based on the number of AAA domain, the members of the AAA gene family can be classified into two types: Type I contains only a single copy of AAA domain and Type II contains two copies of AAA domain termed D1 and D2, respectively (Wang et al. 2004). The AAA domain comprises the conserved motifs (Walker A and Walker B) for ATP binding and hydrolysis (Wang et al. 2003a, b; Zhang et al. 2000). The N domain and C tail are structurally flexible and mainly involved in selecting and/or processing cofactors/substrates, but they could also modulate ATPase activity via either posttranslational modification or protein–protein interaction (Ye 2006; Jentsch and Rumpf 2007; Meyer et al. 2012). In the case of Type II family members, the conformation of the N-domain in relation to the D1-D2 hexamer is directly linked to ATP hydrolysis and that the C-terminal region is required for the hexamer stability (Niwa et al. 2012).
The physiological and biochemical functions of CDC48/p97 protein are mainly manifested by the activity of AAA-ATPase (Peters et al. 1990). It promotes the assembly and disassembly of proteins by using the ATPase activity, and ultimately achieves diverse cellular functions (Xia et al. 2016). AAA proteins exist in both monomeric and oligomeric forms, and only oligomeric form that mostly oligomerizes into hexameric, ring-like structures that act upon their substrates (Dalal and Hanson 2001; Niwa et al. 2012; Rockel et al. 2002; Rouiller et al. 2002; Zhang et al. 2000). CDC48/p97, an important member of the eukaryotic Type II AAA ATPase, is highly conserved in molecular evolution and involved in the regulation of diverse cellular activities (Ballar et al. 2011; Bègue et al. 2016; Franz et al. 2011; Mouysset et al. 2008). It has been shown that mutations in CDC48/p97 would result in a wide range of abnormal phenotypes including lethality of an organism. The substitution of S565G in CDC48 causes altered cold sensitivity and cell apoptosis in S. cerevisiae (Madeo et al. 1997). Overexpression of zebrafish CDC48 promotes cell proliferation and increases DNA synthesis while the expression of a mutant molecule with a tyrosine-805 to alanine substitution at the C-terminal phosphorylation site inhibits cell proliferation and induces apoptosis at low temperatures (Imamura et al. 2003). Overexpression of Trypanosome brucei VCP-1, a CDC48/p97 homologue with a D2 mutation, could completely terminate the cell cycle, thus leading to growth arrest and apoptosis (Lamb et al. 2001). Mutations and RNAi of CDC48 have been found to cause embryonic lethal, abnormal disassembly of spindles, insufficient DNA synthesis and endoplasmic reticulum-associated degradation of proteins in X. laevis and C. elegans (Cao et al. 2003; Mouysset et al. 2008; Ballar et al. 2011). In mammals, CDC48/p97 regulates the process of apoptosis, especially in the pathogenesis of myopathy associated with Paget’s disease of bone and frontotemporal dementia (Haubenberger et al. 2005; Guinto et al. 2007). In human, a point mutation in p97 causes retarded cell proliferation and apoptosis of human B-lymphocytes (Shirogane et al. 1999). In rice, both a single base substitution in OsCDC48 and RNAi of OsCDC48 result in premature senescence and plant death (Huang et al. 2016). Although the mechanism of CDC48-mediated cell death remains to be further elucidated, it is believed that the diverse CDC48 functions depend largely on its cofactors such as the serine/threonine kinase Pim-1 (Shirogane et al. 1999) and the deubiquitinating enzyme ATX-3 (Kuhlbrodt et al. 2011). Thus, identification of cofactors is crucial for the understanding of CDC48-mediated cellular processes.
The rice genome (Oryza sativa L. Japonica. cv. Nipponbare) harbors 29 members of AAA proteins including CDC48 (Os03g0151800). However, very little is known about the roles of OsCDC48 in rice growth, development and senescence. We previously identified a rice premature senescence and death 128 (psd128) mutant resulting from a point mutation of OsCDC48, and an OsCDC48 homologue termed as OsCDC48E (Huang et al. 2016). In this study, we found that OsCDC48E knockout plants exhibited similar behavior to psd128 with premature senescence and death phenotype. The interaction of OsCDC48 with OsCDC48E (Os10g0442600) was necessary for rice plant survival. Removal of the C-terminus of OsCDC48 decreased its ATPase activity, caused the altered expression of cell cycle-related genes, changed the percentage of cells in G1 and G2/M phases, and abolished the interaction between OsCDC48 itself and between OsCDC48 and OsCDC48E, respectively. Overexpression of OsCDC48–psd128 resulted in differential expression of AAA-ATPase associated genes leading to increased total ATPase activity, accumulation of reactive oxygen species and decreased plant tiller numbers while overexpression of OsCDC48 also resulted in differential expression of AAA-ATPase associated genes leading to increased total ATPase activity, but increased plant tiller numbers and grain yield, indicating its potential utilization for yield improvement. Our results demonstrated that the C-terminal region of OsCDC48 was essential for maintaining the full ATPase activity and OsCDC48/48E complex was likely functioning as heteromultimers to regulate cellular processes and plant survival in rice.
Materials and methods
Plant materials and growth conditions
The premature senescence and death 128 (psd128) mutant was obtained from a mutant population generated by ethyl methane sulfonate (EMS) treatment of IR64, an elite indica rice (Oryza sativa L.) cultivar (Huang et al. 2016). The wild type IR64 (WT), psd128 and three backcross F3 lines (psd128/IR64//IR64) with the mutant phenotype were planted in the paddy field under regular water, fertilizer and pest management in the summer of 2017 at the China National Rice Research Institute (CNRRI) in Fuyang, Hangzhou, China. Transgenic plants including overexpression and CRISPR/Cas9-mediated knockout plants were grown in the greenhouse at CNRRI.
Vector construction
For overexpression analysis, the full-length complementary DNAs (cDNAs) of OsCDC48 and OsCDC48–psd128 were amplified, respectively, and the PCR products were then cloned into the binary vector pCambia1305.1-GFP using the Trelief™ SoSoo Cloning Kit (Tsingke, Hangzhou, China) to generate two new constructs pd35S::OsCDC48-GFP and pd35S::psd128-GFP. The pd35S::OsCDC48-GFP and pd35S::psd128-GFP constructs were introduced into the calli generated from the mature embryo of WT, respectively. To knock out OsCDC48E, the targeted deletion vector was constructed following the CRISPR/Cas9 system (Wang et al. 2015) using the target sequence, 5′-CAGGCCTGACATCATAGATC-3′, and introduced into the calli induced from the mature embryo of WT via Agrobacterium tumefaciens-mediated transformation (Hiei and Komari 2008).
Histochemical analysis, H2O2 and malonaldehyde and pigment level determination
Young leaves from WT and overexpression plants at the tillering stage were used for 3,3′-diaminobenzidine (DAB), tryphan blue staining and H2O2 and malonaldehyde (MDA) content determination. The DAB assay was carried out according to method described by Thordal-Christensen et al. (1997). Tryphan blue staining was carried following the method described by Yin et al. (2000). The H2O2 and MDA contents were determined using the method described by Moradi and Ismail (2007). Chlorophyll (Chl) a and Chl b contents were measured using young leaves from WT and CRIPSR-Cas9 knockout plant Cr9-2# at the tillering stage according to the method described by Arnon (1949) while the carotenoid contents from the same leaves were determined following the method described by Wellburn (1994). The means from three replicates were used for analysis.
Flow cytometric analysis
To prepare suspension cells, the 3-day-old shoots of WT and psd128 after germination were soaked in 500 µl 2 µg/ml DAPI solution (DAPI, Beckman, NPE 731085) in a 1.5 ml Eppendorf tube on ice, then cut into pieces and chopped with a sharp razor blade. After filtering the slurry through a 40 µm nylon filter (FALCON 352340), the suspension of nuclei was loaded into a Beckman Moflo-XDP (Beckman Coulter, Inc., CA, USA) for flow cytometric analysis as described by Galbraith et al. (1983), and the ploidy of approximate 10,000 nuclei was recorded for each test. The numbers of diploid and tetraploid nuclei were recorded, and the relative proportions of G1, S, and G2/M cells were calculated using the Summit 2.0 software (Beckman Coulter, USA).
Gene expression analysis
Total RNA was extracted from various organs (flag leaf, flag leaf sheath, culm, root, panicle) of WT and psd128 using Trizol reagent (Invitrogen, Life Technologies, Carlsbad, CA, USA) following the manufacturer’s protocol. The first-strand cDNA was synthesized from DNaseI-treated RNA with an oligo (dT)18 primer in a 20 µL reaction using the ReverTra Ace qPCR Master Mix kit (Toyobo, Tokyo, Japan) following the manufacturer’s protocol. qRT-PCR was conducted using SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) Kit and performed on a Thermal Cycle Dice® Real Time System (TaKaRa, Dalian, China). The rice ubiquitin gene Os03g0234200 was used as an internal control. For spatial and temporal expression analysis, total RNA samples were prepared from the flag leaves, flag leaf sheaths, culms, roots, and panicles of WT and psd128 at the heading stage. For ATPase-related gene expression analysis, total RNA samples were prepared from 15 day-old seedlings of WT and psd128. For cell cycle-related gene expression analysis, total RNA samples were prepared from 3 day-old shoots. In addition, total RNA samples were prepared for CDC48/48E expression analysis from the leaves of CRISPR/Cas9 knockout plants at the tillering stage. All primers used for qRT-PCR are listed in Supplementary Table S1. All assays were repeated at least three times, and the means were used for analysis. The 2− ΔΔCt method (Schmittgen and Livak 2008) was used to calculate relative transcript abundances.
Transcriptome analysis
To eliminate other possible mutations in the mutant, psd128 was first backcrossed to WT. The F1 plants were then backcrossed to WT again to generate BC2F1, which was selfed to generate BC2F2 and BC2F3. Three biological repeats with three siblings/repeat of BC2F3 were used for total RNA extraction with TRIzol reagent (Invitrogen). High throughput RNA sequencing (RNA) was performed by Novogene. Library construction was performed according to Illumina instructions and sequenced on a HiSeq 2000 sequencer. All paired-end reads were mapped to the rice cv. Nipponbare genome using TopHat2 (Trapnell et al. 2009). Expression levels were calculated using the reads per kb permillion reads method (Mortazavi et al. 2008). The DESeq R package (1.10.1) was used for analysis of differentially expression genes (DEGs) between WT and BC2F3. The DEGs were filtered for a corrected P ≤ 0.005. The clustered genes were assigned to biological process categories based on GO analysis using the Web tool DAVID Bioinformatics Resources 6.7 (http://david.abcc.ncifcrf.gov/home.jsp). Significantly enriched GO terms for the DEGs compared with the genomic background were identified using a hypergeometric test (P ≤ 0.05). KEGG pathway-based analysis was performed using the blastall program against the KEGG database (http://www.genome.jp/kegg). Significantly enriched metabolic pathways or signal transduction pathways for the DEGs were identified by pathway enrichment analysis (P ≤ 0.05; Kanehisa et al. 2008).
Subcellular localization
The full-length CDSs of OsCDC48, OsCDC48E and OsCDC48–PSD128 were amplified and cloned into PAN580 to generate three transient expression vectors p35S-CDC48-GFP, and p35S-CDC48E-GFP, p35S-CDC48-PSD128-GFP, respectively. The nuclear marker mCherry-D53 and the cytoplasmic marker mCherry-TAD1 were amplified and cloned into 163-mCherry (Xu et al. 2012; Zhou et al. 2013). The vectors p35S-CDC48-GFP and p35S-CDC48E-GFP were respectively co-transformed with mCherry-D53 and mCherry-TAD1 into the rice protoplasts which were prepared from the stem and leaf sheath of 15-day-old rice seedlings according to the method described by Chen et al. (2006). In addition, p35S-CDC48-GFP and p35S-CDC48-PSD128-GFP were introduced into the protoplasts using the same method, respectively (Chen et al. 2006).The GFP fluorescence was observed by a Zeiss Ism710 confocal laser scanning microscope (Carl Zeiss, Inc., Jena, Germany).
Protein purification and ATPase activity assay
For the recombinant protein expression in Escherichia coli, the CDS of OsCDC48, OsCDC48–PSD128 and OsCDC48E were amplified and cloned into pET28a (Merck, Darmstadt, Germany). In addition, the CDS of OsCDC48 and OsCDC48–PSD128 were also cloned into pGEX-4T-1 (GE Healthcare, Chicago, USA). Recombinant proteins were induced in E. coli strain Rossetta, and purified by Glutathione-Sepharose Resin Protein Purification Kit and 6 × His-Tagged Protein Purification Kit (CWBIO, Beijing, China) according to the manufacture’s instruction, respectively. Both fresh leaf tissues from 15 day-old seedlings of WT, psd128 and overexpression transgenic T1 lines and recombinant protein (His-tag) were used to measure ATPase activity by the ATPase Test Kit according to the manufacture’s instruction (Jiancheng Bioengineering Institute, Nanjing, China).
Western blot assay
For immunoblot analysis, the recombinant proteins (His-tag) were separated on 10% SDS–PAGE gel. Then the target protein bands were sequentially detected with primary antibody anti-His (Cat: CW0083, CWBIO, Beijing, China, 1:5000 dilution) at 4 °C overnight, and secondary antibody with a HRP (Cat: 33101ES60, YEASEN, Shanghai, China, 1:1000 dilution) for 1 h, the immunoblot signal was detected using Supersignal West Pico Chemiluminescent Substrate (Thermo, Waltham, USA) and visualized by the ChemDocTM Touch Imaging system (Bio-Rad, CA, USA).
Yeast two-hybrid and in vitro pull-down assay
Yeast two-hybrid assays were performed with the Y2H Gold-Gal4 system (Clontech, http://www.clontech.com). DNA fragments containing the full coding sequences of OsCDC48, OsCDC48E and OsCDC48–psd128 genes were inserted into the pGBKT7 and pGADT7 vectors to from the bait and prey constructs, respectively. The bait and prey constructs were transformed into yeast strain Y2H Gold according to the manufacturer’s instruction (Clontech, http://www.clontech.com). The yeast cells were cultured on SD/-Trp-Leu or SD/-Trp-Leu-His-Ade medium containing X-ɑ-gal and Aureobasidin A (AbA) at 30 °C in darkness for 3 days. The pull-down assay was conducted as follows: 50 µL equilibrated Glutathione High Capacity Magnetic Agarose Beads (Sigma, St Louis, USA) was mixed with 500 µg of each recombinant protein in 600 µL pull-down buffer (50 Mm Tris–HCl pH = 7.5, 5% glycerol, 1 mM EDTA, 1 mM DTT, 1 Mm PMSF, 0.01% Nonidet P-40, 150 mM KCl) under 4 °C for 2 h. The bound proteins together with the beads were collected by a magnetic shelf (Invitrogen, Carlsbad, USA), washed with pull-down buffer twice, eluted with 50 µL 1 × PBS and immune detected by GST (Cat: CW0085, CWBIO, Beijing, China), HIS (Cat: CW0083, CWBIO, Beijing, China) antibodies respectively.
BiFC assay
For the BiFC assay, the full length CDSs of OsCDC48, OsCDC48E, and OsCDC48–psd128 genes were cloned into pCAMBIA1300S-YN and pCAMBIA2300S-YC to form the nYFP-protein and cYFP-protein constructs, respectively. The constructs then were transformed into rice protoplasts according to the protocols described previously by Zhang et al. (2011). In addition, the constructs also were transiently expressed in tobacco leaves following the method described by Waadt and Kudla (2008). The BiFC assay was performed as described previously (Waadt and Kudla 2008). A confocal laser scanning microscope (Zeiss LSM710) was used to detect YFP fluorescent signals after 48 h post-transfection.
Results
OsCDC48E belongs to the AAA-ATPase family protein
We previously isolated OsCDC48 in a study of the rice premature senescence and death 128 (psd128) mutant (Huang et al. 2016). A single base substitution at position C2347T leading to a premature stop mutation (OsCDC48–PSD128) resulting in a putative truncated protein (OsCDC48–PSD128) lacking of 27 amino acid residues at the C-terminus, is responsible for the premature senescence and death phenotype of psd128. We also identified an unknown function OsCDC48 homologue, OsCDC48E, which is localized to chromosome 10 and shares 97% identity to OsCDC48 at the amino acid level (Huang et al. 2016). In the present study, we showed that the cDNA of OsCDC48E (accession number, MK292711) compromised of 2882 bp including a 145 bp 5′-UTR, a 310 bp 3′-UTR and a 2427 bp CDS. Similar to OsCDC48, OsCDC48E has 9 exons and 8 introns, and encodes a putative AAA-ATPase with 808 amino acid (aa) residues with a predicted molecular mass of 96.96 kDa. Similar to OsCDC48, the putative OsCDC48E protein has a typical N terminus (1–199 aa), a C terminus (772–808 aa) and two AAA-ATPase domains (D1 from 213 to 466 aa and D2 from 486 to 771 aa) (Fig. S1). Each of the AAA-ATPase domains contains the Walker A and Walker B motifs (Fig. S1). The results indicated that OsCDC48E belongs to the AAA-ATPase family protein.
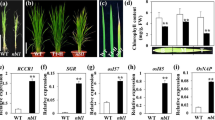
OsCDC48E is constitutively expressed
OsCDC48 is constitutively expressed in the tissues tested (Huang et al. 2016). To determine the expression pattern of OsCDC48E, Real-time PCR was carried out and the results revealed that OsCDC48E had a similar expression pattern to OsCDC48, and was constitutively expressed in different tissues including the roots, culms of the second internode, flag leaf sheaths, flag leaves and panicles at the heading stage (Fig. 1). The highest expression levels of OsCDC48 and OsCDC48E were detected in the flag leaves while the lowest levels of OsCDC48 and OsCDC48E were detected in the culms in IR64 (WT). In contrast, the highest expression levels of OsCDC48 and OsCDC48E were detected in the flag leaf sheaths while the lowest levels of OsCDC48 and OsCDC48E were detected in the roots and flag leaves in psd128 (Fig. 1). These results suggested that the expression patterns of OsCDC48 and OsCDC48E were similar both in psd128 and WT at the heading stage.
OsCDC48E and OsCDC48 localize to nucleus and cytoplasm
In the previous study, we localized OsCDC48-GFP to the nucleus and cytoplasm in the mesophyll cells of Nicotiana tabacum (Huang et al. 2016). To confirm the subcellular localization of OsCDC48-GFP in rice nuclei, the transient vector p35S-CDC48-GFP was co-transformed with the nuclear marker mCherry-D53 using rice protoplasts. The results showed that both p35S-CDC48-GFP and mCherry-D53 localized to the rice nucleus, indicating that OsCDC48-GFP was indeed localized to the nucleus (Fig. 2a–d). To confirm the subcellular localization of OsCDC48-GFP in the cytoplasm in rice, the transient vector p35S-CDC48-GFP was co-transformed with the cytoplasmic marker mCherry-TAD1. The results showed that both p35S-CDC48-GFP and mCherry-TAD1 localized to the rice cytoplasm, indicating that OsCDC48-GFP was indeed localized to the cytoplasm as well (Fig. 2e–h). To determine the subcellular location of OsCDC48E in rice, similar co-transformation assays with mCherry-D53 and mCherry-TAD1 were carried out using rice protoplasts, and the results showed that OsCDC48E localized both to the nucleus (Fig. 2i–l) and cytoplasm (Fig. 2m–p). To determine the effect of C-terminal deletion on subcellular location, the transient expression vector pCDC48-PSD128-GFP was introduced into rice protoplasts, and the results showed that OsCDC48–PSD128-GFP localized to the nucleus and cytoplasm similar to OsCDC48-GFP and OsCDC48E-GFP (Fig. S2), indicating that the deletion of 27 aa residues at the C-terminus did not affect the subcellular location of OsCDC48. Taken together, our results confirmed that both OsCDC48 and OsCDC48E localized to the nucleus and cytoplasm.
Subcellular localization of OsCDC48 and OsCDC48E in rice protoplast. a–d Subcellular localization of OsCDC48-GFP. The nuclear marker OsD53 fused with mCherry was used as a positive control. Bar = 10 µm; e–h subcellular localization of OsCDC48-GFP. The cytoplasmic marker OsTAD1 fused with mCherry was used as a positive control. Bar = 10 µm; i–l subcellular localization of OsCDC48E-GFP. The nuclear marker OsD53 fused with mCherry was used as a positive control. Bar = 10 µm; m–p subcellular localization of CDC48E-GFP. The cytoplasmic marker OsTAD1 fused with mCherry was used as a positive control. Bar = 5 µm
C-terminus of OsCDC48 is essential for its ATPase activity
The diverse cellular functions of CDC48/p97 protein are mainly manifested by the activity of AAA-ATPase (Peters et al. 1990). To determine whether OsCDC48 and OsCDC48E possess the ATPase activity, we first measured the total ATPase activity in 2 week-old seedling leaves of WT and psd128 (Fig. 3a), and the results indicated that the total ATPase activity in psd128 was significantly higher than that of WT (Fig. 3b). The results implied that other members of AAA-ATPases might contribute to the increased level of total ATPase activity in psd128. Therefore, we selected seven ATPase-related genes and determined their expression levels by qRT-PCR analysis. As expected, the expression levels of OsAAA-ATPase1, OsAAA-ATPase4, OsAAA-ATPase5 OsAAA-ATPase6 and OsAAA-ATPase7 in psd128 were significantly higher than those of WT whereas the expression levels of OsAAA-ATPase2 and OsAAA-ATPase3 in the mutant were similar to those of WT (Fig. 3c). The results clearly demonstrated that the mutation of OsCDC48 induced higher expression levels of a number of other AAA-ATPase genes probably for a compensation of OsCDC48 enzymatic activity. To measure the enzymatic activity of OsCDC48, OsCDC48–PSD128 as well as OsCDC48E, the corresponding proteins encoded by OsCDC48, OsCDC48E and psd128 were expressed in the prokaryotic expression system, purified and identified using anti-6 × His antibody by western blot (Fig. S3), and used to measure their ATPase activities. The result showed that OsCDC48, OsCDC48E and OsCDC84-PSD128 all possessed ATPase activity. However, the activity of OsCDC48–PSD128 was significantly lower than that of the wild-type OsCDC48 (Fig. 3d, e) while the activity of OsCDC48E was similar to that of OsCDC48 (Fig. 3d, e). All these results demonstrated that OsCDC48, OsCDC48E and OsCDC48–PSD128 all possessed ATPase activity and the C-terminus of OsCDC48 was essential for the full ATPase activity of OsCDC48.
ATPase activity and expression of ATPase-related genes. a Two week-old seedlings of WT and psd128 were used for analysis. Bar = 4.5 cm; b total ATPase activities of WT and psd128. Values are means ± SD from three biological replicates. Significance at **P ≤ 0.01 by Student’s t-test; c expression of AAA-ATPase related genes. Values are means ± SD from three biological replicates. Asterisks indicate significance by Student’s t-test (*P ≤ 0.05; **P ≤ 0.01). OsCDC48 (Os03g0151800), OsCDC48E (Os10g0442600), OsAAA-ATPase1 (Os01g0297200), OsAAA-ATPase2 (Os07g0192800), OsAAA-ATPase3 (Os07g0517800), OsAAA-ATPase4 (Os12g0431100), OsAAA-ATPase5 (Os12g0468000), OsAAA-ATPase6 (Os12g0639200) and OsAAA-ATPase7 (Os05g0588850); d purification of OsCDC48, OsCDC48E and OsCDC48–PSD128 (red arrows indicate the protein bands). The purified fraction (1 µg) was run on 10% SDS–PAGE and visualized by CBB staining. M, Protein Marker; BSA, Protein standard (20 µg); e ATPase activities of the purified OsCDC48, OsCDC48E and OsCDC48–PSD128, respectively, Values are means ± SD from three biological replicates. Different letter indicates significance at P ≤ 0.01 by Student’s t-test
Overexpression of OsCDC48 increases total ATPase activity and promotes plant development
To investigate the biological effects of OsCDC48, we generated transgenic plants overexpressing the wild-type OsCDC48 and the mutant (OsCDC48–psd128) alleles under the control of double 35S promoters in WT background. A total of 6 OsCDC48 and 7 OsCDC48–psd128 T0 overexpression plants were obtained respectively. Two independent plants each representing the WT (L2 and L4) and mutant type (L3 and L5) alleles were chosen for further study respectively (Fig. 4a). We found that the expression levels of OsCDC48 and OsCDC48–psd128 in the T0 plants were approximately 2- and 8- fold higher than those of WT at the heading stage, respectively (Fig. 4b). Notably, the transgenic plants overexpressing OsCDC48 showed an exaggerated phenotype with significantly decreased 1000-grain weight, increased tiller number and grain yield compared with WT (Fig. 4a, c; Table S2). In contrast, the transgenic plants overexpressing OsCDC48–psd128 showed a minimized phenotype with significantly reduced plant height, decreased tiller numbers and premature senescence similar to psd128 compared to WT (Fig. 4a, c). The differential phenotypic performances of these transgenic plants were correlated with the expression level of the transgenes (Fig. 4b, c). Moreover, we found that the expression levels of OsCDC48 and OsCDC48–psd128 in the respective transgenic T1 lines were 2.7 (L2), 2.1(L4), 6.9 (L3) and 11.1 (L5) fold higher than those of WT at the seedling stage, respectively (Fig. 4d). Notably, all the T1 lines overexpressing both OsCDC48–psd128 and OsCDC48 at the seedling stage showed significantly increased total ATPase activity compared with WT (Fig. 4e). The results implied that other members of AAA-ATPases might also contribute to the increased level of total ATPase activity in overexpression T1 lines. Therefore, we further determined their expression levels of 7 ATPase-related genes in L5 (OX-OsCDC48–PSD128) and L2 (OX-OsCDC48) by qRT-PCR, respectively. The results showed that the expression levels of OsAAA-ATPase1, OsAAA-ATPase2, OsAAA-ATPase4, OsAAA-ATPase5 OsAAA-ATPase6 and OsAAA-ATPase7 in L5 were significantly higher than those of WT whereas the expression levels of OsCDC48E and OsAAA-ATPase3 in L5 were significantly lower than those of WT (Fig. 4f). In contrast, the expression levels of OsAAA-ATPase3, OsAAA-ATPase4, OsAAA-ATPase5, OsAAA-ATPase6 and OsAAA-ATPase7 in L2 were significantly lower than those of WT whereas the expression levels of OsCDC48 and OsCDC48E in L2 were significantly higher than those of WT, and the expression levels of OsAAA-ATPase1, OsAAA-ATPase2 were similar to those of WT (Fig. 4f). These results demonstrated that increased total ATPase activity was contributed by other members of ATPase-related genes in L5, while increased total ATPase activity in L2 was contributed by OsCDC48 and OsCDC48E. Excessive amount of ROS causes premature senescence and cell injury/death (Davletova et al. 2005). To investigate the role of OsCDC48 in leaf senescence, we carried out DAB and tryphan blue staining on two transgenic T0 plants overexpressing OsCDC48–PSD128 (L5) and OsCDC48 (L2) at the heading stage. The results showed that little brown precipitate and blue stains were detected on the leaves of L2, however, an apparent increased amount of brown precipitate and blue stains were detected on the leaves of L5, indicating that overexpression of the mutation allele induced H2O2 accumulation and lead to cell death (Fig. 4g, h). We then further measured the concentrations of H2O2 and MDA, the results exhibited that concentrations of both H2O2 and MDA were significantly higher in L5 leaves than those of L2 (Fig. 4i, j). Again, the severity of ROS accumulation and leaf senescence phenotype in the transgenic plants were correlated with the expression levels of the transgenes. To further explore the potential mechanism associated with premature leaf senescence in L5, we measured the expression of two senescence indicators. The results showed that the expression levels of Osh36 and OsI57 were apparently upregulated in L5 as well as in L3 while their expression levels were significantly downregulated in L2 as well as in L4 (Fig. S4A, B). Taken together, our results demonstrated that overexpression of OsCDC48–psd128 induced ROS accumulation, premature leaf senescence and reduced plant tiller numbers and overexpression of OsCDC48 delayed senescence and increased the grain yield mainly by producing more effective plant tillers.
Comparison of OsCDC48 and OsCDC48–psd128 overexpression transgenic lines. a Phenotype of WT, overexpression T0 lines carrying pd35s::OsCDC48-GFP (L2 and L4) and pd35s::psd128-GFP (L3 and L5) at the heading stage. Bar = 20 cm; b relative expression of OsCDC48 in overexpression T0 lines at the heading stage. Values are means ± SD from three biological replicates. Significance at **P ≤ 0.01 by Student’s t-test; c tiller number of T0 lines. Significance at **P ≤ 0.01 by Student’s t-test; d relative expression of OsCDC48 in overexpression T1 lines of 15-day-old seedlings. Values are means ± SD from three biological replicates. Significance at **P ≤ 0.01 by Student’s t-test; e total ATPase activity of overexpression T1 lines of 15-day-old seedlings. Significance at **P ≤ 0.01 by Student’s t-test; f expression of AAA-ATPase related genes in overexpression T1 lines of 15-day-old seedlings. Values are means ± SD from three biological replicates. Asterisks indicate significance by Student’s t-test (*P ≤ 0.05; **P ≤ 0.01). OsCDC48 (Os03g0151800), OsCDC48E (Os10g0442600), OsAAA-ATPase1 (Os01g0297200), OsAAA-ATPase2 (Os07g0192800), OsAAA-ATPase3 (Os07g0517800), OsAAA-ATPase4 (Os12g0431100), OsAAA-ATPase5 (Os12g0468000), OsAAA-ATPase6 (Os12g0639200) and OsAAA-ATPase7 (Os05g0588850); g DAB staining of WT, L2 and L5 T0 leaves at the heading stage (Left, before staining; Right, after staining); h Trypan blue staining of WT, L2 and L5 T0 leaves at the heading stage (Left, before staining; right, after staining); i H2O2 contents of WT, L2 and L5 T0 plants at the heading stage; j MDA contents of WT, L2 and L5 T0 plants at the heading stage. Data are the means ± SD from three replicates. Different letters on the error bars indicate significance at P ≤ 0.01 by Duncan’s test. FW fresh weight
C terminal mutation of OsCDC48 affects cell cycle progression
The yeast CDC48 mutant is defective in the cell cycle (Moir et al. 1982). To determine whether the mutation of OsCDC48 affected the progression of cell cycle, we investigated the cell status in the cell cycle using shoot suspension cells by flow cytometry. The results revealed that the number of G1 cells in psd128 was significantly lower than that of WT, and the number of G2/M cells in psd128 was significantly higher than that of WT while the number of cells in S phase was similar between psd128 and WT (Fig. 5a–c). In addition, the cell size was similar between psd128 and WT while the cell number of internode VI in psd128 was significantly lower than that of WT at the heading stage (Fig. S5A–C). The results demonstrated that OsCDC48 was involved in regulation of cell cycle progression and the mutation affected both the interphase and cell division. We then determined the expression levels of 7 cell cycle-related genes in WT and psd128 plants using qRT-PCR. Our results showed that the expression levels of G1-related CDKA1 and CAK1A in psd128 were significantly down-regulated by 30.28 and 33.06%, respectively, and the expression levels of G1-related MCM5 and CYCT1 in psd128 were up-regulated by 5.55-fold and 86.51%, respectively, compared with WT (Fig. 5d). Furthermore, the expression levels of G2-related CYCA2.2, CYCA2.3 and CYCB2.2 in psd128 were significantly up-regulated by 1.22-fold, 2.07-fold and 71.97%, respectively, compared to WT (Fig. 5d). In addition, we further determined the expression of cell cycle-related genes in overexpressing T1 lines. Interestingly, the results exhibited that the expression levels of most these genes were significantly altered but with different patterns compared with psd128 (Fig. S4C–I). Taken together, we concluded that OsCDC48 played a critical role in the cell cycle by regulating the expression of a number of cell cycle-associated genes in rice.
Flow cytometric and cell cycle-related gene expression analysis. a, b Flow karyotype histogram of WT (a) and psd128 (b), the x-axis represents the relative fluorescence intensity, the y-axis represents the number of cells, and the peaks under the open boxes represent the phases of the cells; c Percentage of cells at different phases from 3 day-old shoots in WT and psd128. Significance at **P ≤ 0.01 by Student’s t-test; d relative expression levels of cell cycle related genes in 3 day-old shoots of WT and psd128. Values are means ± SD from three biological replicates. Asterisks indicate significance by Student’s t-test (*P ≤ 0.05, **P ≤ 0.01)
C-terminal deletion affects the expression of diverse genes genome-wide
To investigate the potential roles and mechanisms associated with OsCDC48, we carried out high throughput mRNA sequencing (RNA-Seq). The cDNA libraries were prepared from the leaves of 3 WT individual plants and three BC2F3–psd128 lines (mutant-like) derived from psd128/IR64//IR64, respectively. Our results revealed that a total of 374 differentially expressed genes (DEGs) were identified between BC2F3–psd128 and WT (Supplementary Data S1, total). Among them, 232 genes were up-regulated and 142 genes were down-regulated in BC2F3–psd128. The cell cycle-associated genes including CYCA2.3 (Os01g0233500), CYCT1 (Os02g0438200), MCM5 (Os02g0797400), CYCB2.2 (Os06g0726800) and CYCA2.2 (Os12g0502300) were significantly up-regulated by 2.40, 1.47, 3.78, 1.32 and 1.52 fold, respectively (Supplementary Data S1, up-regulated). CDKA1 (Os03g0118400) and CAK1A (Os06g0334400) were significantly down-regulated by 1.11 and 1.02 fold, respectively (Supplementary Data S1, down-regulated). The results were similar to those of qRT-PCR. In addition, a total of 615 genes were expressed only in WT, and 2190 genes covering a set of AAA-ATPase-encoding genes were expressed only in BC2F3–psd128 (Table S4). Furthermore, the AAA-ATPase encoding genes including OsAAA-ATPase1 (Os01g0297200), OsAAA-ATPase4 (Os12g0431100), OsAAA-ATPase5 (Os12g046800), OsAAA-ATPase6 (Os12g0639200) and OsAAA-ATPase7 (Os05g0588850) were significantly up-regulated by 1.13, 2.90, 2.62, 1.41 and 3.92 fold in BC2F3–psd128, respectively (Supplementary Data S1, up-regulated). OsCDC48 (Os03g0151800) and OsCDC48E (Os10g0442600) were significantly down-regulated by 1.38 and 1.51 fold in BC2F3–psd128, respectively (Supplementary Data S1, down-regulated). The expression levels of OsAAA-ATPase2 (Os07g0192800), OsAAA-ATPase3 (Os07g0517800) and other AAA-ATPase associated genes were similar between WT and BC2F3–psd128 (Supplementary Table 4). Again, the results were similar to those of qRT-PCR. Taken together, the C terminal mutation affected the cell cycle progression and the global AAA-ATPase activity.
We then performed Gene Ontology (GO) analysis to classify the functions of the 374 DEGs identified. The GO term enrichment indicated that these DEGs could be classified into 74 and 44 GO terms under three biological processes with P ≤ 0.05 and ≤ 0.01, respectively (Supplementary Data S2, total). Among the 44 highly significant GO terms, 5 terms belong to Functional process, 16 terms belong to Biosynthetic process, and 23 terms belong to Component process (Supplementary Data S2, total). For the 232 up-regulated genes, a total of 40 GO terms were assigned, and 13 out 40 terms were highly significant terms such as genes that were associated with cell wall biogenesis/organization and cell division (Supplementary Data S2, up-regulated). For the 142 down-regulated DEGs, a total of 89 GO terms were assigned, and 66 out 89 terms were highly significant terms covering genes that were associated with chloroplast biogenesis/development, chlorophyll biosynthesis and photosynthesis (Supplementary Data S2, down-regulated). The results further suggested that OsCDC48 was involved in the cell cycle and chloroplast development in rice.
To further explore the biological pathways in which the mutation may be involved, we performed KEGG enrichment analysis for the 374 DEGs between WT and BC2F2–psd128. The results showed that all the DEGs were classified into 8 pathways (Supplementary Data S3, total). The up-regulated 232 DEGs were significantly enriched in 3 pathways including phenylpropanoid biosynthesis, phenylalanine metabolism and glutathione metabolism (Supplementary Data S3, up-regulated), while the 142 down-regulated DEGs were grouped into 6 predominant pathways including photosynthesis-antenna proteins, porphyrin and chlorophyll metabolism, thiamine metabolism, metabolic pathway, photosynthesis, biosynthesis of secondary metabolites (Supplementary Data S3, down-regulated). The results suggested that the C terminal mutation of OsCDC48 affected multiple pathways including amino acid metabolism probably required for cellular protection and photosynthesis-related metabolism.
C-terminus is required for CDC48/48, CDC48E/48E and CDC48/48E interaction in vivo
CDC48/p97, one of the most abundant proteins in eukaryotic cells, is a molecular chaperone evolutionally conserved in plants, yeasts and animals (Wolf and Stolz 2012). It functions as a hexamer although the monomeric form exists (Bègue et al. 2016). To evaluate the biochemical function of OsCDC48/48E and the effect of C-terminal deletion on OsCDC48 function, we first performed a yeast two-hybrid (Y2H) assay with the full-length versions of OsCDC48, OsCDC48E and the truncated OsCDC48–PSD128. Our results revealed that OsCDC48 interacted with OsCDC48 itself, and OsCDC48E interacted with OsCDC48E itself as well. Interestingly, OsCDC48 could interact with OsCDC48E. However, the truncated OsCDC48–PSD128 interacted neither with OsCDC48–PSD128 itself nor with OsCDC48 and OsCDC48E, respectively (Fig. 6a–c). Then, we carried out a pull-down assay to validate the interactions among OsCDC48, OsCDC48E and OsCDC48–PSD128 in vitro. The results revealed that both OsCDC48 and OsCDC48E could interact with itself, and OsCDC48 could interact with OsCDC48E while no interaction was detected between OsCDC48–PSD128 molecules (Fig. 6d). Finally, we performed a bimolecular fluorescence complementation (BiFC) assay in vivo. Consistent with Y2H, the BiFC assay showed that both OsCDC48 and OsCDC48E could interact with itself and each other, while OsCDC48–PSD128 could not interact with OsCDC48, OsCDC48E and itself (Figs. 6e, S6). Taken together, these results indicated that OsCDC48/48E was likely functioning as a hetero-oligomers and the C-terminus played a crucial role for the interaction.
Interaction of OsCDC48 and OsCDC48E in vitro and vivo. a–c Yeast two-hybrid (Y2H) assay. Serial dilutions (10-fold) of yeast cells expressing the indicated proteins were plated onto SD/-LT nonselective medium (SD/-Leu/-Trp) (Left) and SD/-LTHA selective medium (SD/-Leu/-Trp/-His/-Ade) (Right) supplemented with X-α-gal and AureobasidinA (AbA). pGADT7-T (T) /pGBKT7-53 (53) was used as the positive control and pGADT7-T (T)/pGBKT7-laminC (laminC) was used as the negative control, respectively. a Y2H assay shows that OsCDC48 interacts with itself and OsCDC48–PSD128 could not interact with itself; b Y2H assay shows that OsCDC48 interacts with OsCDC48E, and OsCDC48E interacts with itself; c Y2H assay shows that OsCDC48 interacts with OsCDC48E and OsCDC48–PSD128 could not interact with OsCDC48E; d in vitro pull-down assay. The fusion proteins of OsCDC48, OsCDC48–PSD128 and OsCDC48E with a His tag (His-CDC48, His-PSD128 and His-CDC48E) and OsCDC48 and OsCDC48–PSD128 with a GST tag (GST-CDC48, GST-PSD128) were detected by anti-His antibody and anti-GST antibody, respectively; e in vivo bimolecular fluorescence complementation assay shows that OsCDC48 and OsCDC48E interacts with itself, respectively, but OsCDC48–PSD128 does not interact with itself, and OsCDC48 interacts with OsCDC48E in rice protoplast. Bars = 5 µm. YN + YC indicates negative control; YN-OsHAL3 + YC-OsHAL3 indicates positive control
OsCDC48E knockout plants exhibit premature senescence and death phenotype
Having established that OsCDC48E was required for the interaction with OsCDC48, we speculated that OsCDC48E alone must be essential for plant survival in rice. To evaluate this possibility, we knocked out OsCDC48E in the wild-type (IR64) background using the CRISPR/Cas9 approach (Wang et al. 2015). The results showed that a total of 9 positive transformants (T0) were obtained and all of them exhibited dwarfism, premature senescence and death before heading, mimicking the psd128 phenotype although psd128 was able to set a few seeds before completely died (Fig. 7; Table S3). Among the 9 CRISPR-Cas9 knockout plants, Cr9-2# and Cr9-3# displayed both premature senescence phenotype and shorter plant height at the heading and tillering stages, respectively (Fig. 7a, b; Fig S7A). Cr9-2# was a homozygous mutant carrying a T nucleotide insertion in the 8th exon (Fig. 7c, e) while Cr9-3# was a homozygous mutant carrying a 5-bp (CATAG) deletion in the targeted 8th exon (Fig. 7d, e). As expected, the expression levels of OsCDC48E in Cr9-2# and Cr9-3# were significantly reduced compared to WT (Fig. 7f, g). In addition, Cr9-2# exhibited a premature senescence phenotype with significantly reduced levels of photosynthetic pigments (Fig. S7B) similar to psd128 (Huang et al. 2016). Furthermore, the remaining 7 OsCDC48E knock out plants died at a very young seedling stage (data not shown).We hypothesize that OsCDC48 and OsCDC48E are likely to form a heteromultimeric complex to control plant survival by regulating senescence-associated genes and the cell cycle progression in rice.
Phenotypic characterization and expression analysis of OsCDC48E-knock out lines. a Phenotype of WT (left) and knock out plant Cr9-2# (right) at the heading stage. Bar = 5 cm; b phenotype of WT (Left) and knock out plant Cr9-3# (right) at the tillering stage. Bar = 5 cm; c, d structure of OsCDC48E: the empty box indicates 5′UTR, black boxes indicate exons, lines indicate introns and white arrows indicate 3′UTR and the red transverse lines indicate nucleotide mutations in Cr9-2# and Cr9-3#, respectively. The last 3 nucleotides in the black shadow area are PAM sequence (CGG); e mutations in Cr9-2# and Cr9-3#. Cr9-2# is a homozygous mutant carrying a T insertion and Cr9-3# is a homozygous mutant carrying a 5-bp deletion. The sgRNA target sequence is underlined in blue and the PAM motif is highlighted in red letters; fOsCDC48 and OsCDC48E expression levels in WT and Cr9-2# at the heading stage; gOsCDC48 and OsCDC48E expression levels in WT and Cr9-3# at the tillering stage. Values are means ± SD from three biological replicates. Significance at **P ≤ 0.01 by Student’s t-test
Discussion
OsCDC48 involves in cell cycle regulation and is essential for rice growth and survival
The cell cycle plays a crucial role in regulating the growth and development of organisms. Defective in the cell cycle would lead to abnormal cell proliferation and differentiation, division and cell death (Hartwell and Weinert 1989). Previous studies have inspected the biological significance of rice genes including GSN1, DEL1and TAD1, and confirmed that they are involved in the cell cycle progression and ultimately affect the leaf development and senescence, plant height and grain yield (Guo et al. 2018; Leng et al. 2017; Xu et al. 2012). The initiation and establishment of plant branches/tillers is a complex biological process involving a combination of multiple factors (Wang and Li 2011) and is ultimately achieved through the basic biological process of cell division (Shimizu and Mori 1998). In the present study, OsCDC48 appeared to play an important role in the control of leaf senescence, plant development and survival in rice. Overexpression of OsCDC48 delayed leaf senescence and significantly increased the tiller numbers leading to increased grain yield, suggesting that OsCDC48 is a potential genetic factor for rice yield improvement. In contrast, overexpression of OsCDC48–psd128 dramatically reduced the tiller numbers, plant height and caused premature leaf senescence and plant death because of elevated accumulation of H2O2, indicating that H2O2 signaling might participate in the control of tillering and plant height. Meanwhile, the level of MDA, an indicator of cell membrane damage, was highly elevated in the OsCDC48–psd128 overexpression plants which exhibited premature senescence similar to psd128 (Fig. 4d–m). Our results supported the conclusion that MDA and H2O2 over production could promote senescence in plant cells (Brodersen et al. 2002). The leaf senescence and survival of psd128 is likely resulted from abnormal cell cycle progression manifested in G1 and G2/M phases (Fig. 5a–c). Consistent with the flow cytometric results, the expression levels of genes regulating the cell cycle were significant altered in psd128 (Fig. 5d). In addition, histochemical examination showed that the dwarf phenotype of psd128 was caused by the shortened internodes resulting from a marked reduction of cell number in psd128 (Fig. S4), suggesting that cell division was inhibited. Moreover, although the expression of cell cycle-related genes in overexpressing T1 lines was significantly altered while the patterns were somewhat different from those of psd128, further experiments are necessary to be carried out for clarification (Fig. S5). Nevertheless, our results strongly suggest that OsCDC48 controls plant growth, development and survival by regulating the cell cycle progression in rice.
C-terminus of OsCDC48 influences ATPase activity
A typical eukaryotic AAA-ATPase molecular structure contains an N-terminal domain, an AAA domain and a C-terminal domain. The biochemical functions for a broad utility of p97/CDC48 lie in its ATPase activity which is believed to be carried out mainly through the AAA domain by hydrolyzing ATP to provide energy for diverse cellular functions (Peters et al. 1990). The ATPase activity of p97/CDC48 was first demonstrated in Xenopus laevis (Peters et al. 1990). The N-terminal domain and C-terminal of p97/CDC48, are also required for modulating ATPase activity via either posttranslational modification or other pathways (Jentsch and Rumpf 2007; Wang et al. 2004). It has been shown that the N-domain position relative to the D1 ring is linked to ATP hydrolysis ability and removal of the C-terminal region reduces ATPase activity probably by affecting the hexamer stability (Niwa et al. 2012). In the present study, psd128 is defective in the C terminus, thus we focus on whether the ATPase activity is affected with removal of the C-terminal 27 aa residues of OsCDC48 in rice. Interestingly, the total ATPase activities in leaves were significantly increased in psd128 while the purified OsCDC48–PSD128 exhibited a significantly reduced ATPase activity compared to the WT OsCDC48 protein. Increased total ATPase activity was contributed by other members of ATPase-related genes in psd128 and OsCDC48–psd128 overexpression lines. In contrast, increased total ATPase activity in OsCDC48 overexpression lines was resulted from elevated enzymatic activities of OsCDC48 and OsCDC48E (Fig. 4f). This observation supports that the C terminal defection reduces ATPase activity that is likely associated with hexamer stability which requires to be validated in OsCDC48. Additionally, removal of the C terminal region of OsCDC48 induced significantly elevated levels of expression of a set of other members of AAA-ATPase genes in psd128, however, it could not fully compensate for the role by OsCDC48 as the mutant eventually dies at the heading stage (Huang et al. 2016). The rice genome (cv. Nippobare) harbors 29 members of AAA proteins including CDC48. It would be necessary to investigate the effect of the C terminal defect on all the members so as to provide an insight into the link among the members although a number of AAA-ATPases showing similar expression levels between two genotypes were detected by RNA-seq (Table S5). Besides, the C terminal defect influenced the expression of a large number of genes as shown by RNA-seq between the two genotypes, indicating that the stability of the gene network plays a crucial role at the molecular and cellular levels for rice growth, development and survival. Taken together, we conclude that the C-terminal region of OsCDC48 is critical to plant growth, development and survival via controlling the ATPase activity.
OsCDC48/48E hetero-oligomeric complex is essential for rice survival
In eukaryotic cells, p97/CDC48 is a highly abundant hexameric AAA-ATPase that functions as a molecular chaperone in diverse cellular activities. The assembly and disassembly of the hexameric p97/CDC48 complex itself is a dynamic process (Paro et al. 2007). The AAA domain is highly conserved evolutionally while both the N domain and C domain are structurally flexible and able to modulate protein–protein interaction (Jentsch and Rumpf 2007; Wang et al. 2004). The main function of the N-domain is to control the binding of cofactors and ubiquitylated protein substrates (Jentsch and Rumpf 2007; Wang et al. 2004), and the conformation of the N-domain in relation to the D1-D2 hexamer is directly linked to ATP hydrolysis (Niwa et al. 2012). The C domain contains a tyrosine phosphorylation site that is thought critical to multiple p97/CDC48 functions (Wang et al. 2003a, b). Moreover, the C domain is also considered necessary for hexamer stability and full ATPase activity (Niwa et al. 2012). In the present study, the most striking finding is that interaction between OsCDC48 (Os03g0151800) and OsCDC48E (Os10g0442600) was required for its function in the control of leaf senescence, growth, development and survival, indicating that OsCDC48/48E may form and function as a heterohexamer (Arlt et al.1996; Rubin et al. 1998; Turner et al.1999). In contrast to previous report, p97/CDC48 has been thought functioning in the homo-hexameric form (Peters et al. 1990; Zhang et al. 2000). Whether OsCDC48/48E form and act as a heterohexamer is still required to be validate structurally, however, several observations in the present study support this speculation. Firstly, the expression pattern of OsCDC48 and OsCDC48E was highly similar, indicating that they might act together and have similar functions or are involved in the same pathways. Secondly, both OsCDC48, and OsCDC48E are localized to the nucleus and cytoplasm, indicating that they might function in the same cellular sites with similar roles in relevant biological processes. Lastly, OsCDC48E knockout plants exhibit similar phenotype to psd128 especially in terms of premature senescence and plant lethality, indicating an irreplaceable role by OsCDC48E and only presence of both of them could secure the normal growth, development and senescence. Taken together, the results suggest that OsCDC48 and its homologue OsCDC48E might coordinate in function to regulate plant growth and development.
References
Arlt H, Tauer R, Feldmann H, Neupert W, Langer T (1996) The YTA10-12 complex, an AAA protease with chaperone-like activity in the inner membrane of mitochondria. Cell 85:875–885
Arnon DI (1949) Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol 24:1–15
Ballar P, Pabuccuoglu A, Kose FA (2011) Different p97/VCP complexes function in retrotranslocation step of mammalian Er-associated degradation (ERAD). Int J Biochem Cell B 43:613–621
Bègue H, Jeandroz S, Blanchard C, Wendehenne D, Rosnoblet C (2016) Structure and functions of the Chaperone-like p97/CDC48 in plants. Biochim Biophys Acta 1861:3053–3060
Brodersen P, Petersen M, Pike HM et al (2002) Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Gene Dev 16:490–502
Cao K, Nakajima R, Meyer HH, Zheng YX (2003) The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115:355–367
Chen SB, Tao LZ, Zeng LR, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7:417–427
Dalal S, Hanson PI (2001) Membrane traffic: What drives the AAA. motor? Cell 104:5–8
Davletova S, Schlauch K, Coutu J, Mittler R (2005) The Zinc-finger protein Zat12 plays a central role in reactive oxygen and abiotic stress signaling in Arabidopsis. Plant Physiol 139:847–856
Forman MS, Mackenzie IR, Cairns NJ et al (2006) Novel ubiquitin neuropathology in frontotemporal dementia with valosin-containing protein gene mutations. J Neuropath Exp Neur 65:571–581
Franz A, Orth M, Pirson PA et al (2011) CDC-48/p97 coordinates CDT-1 degradation with GINS chromatin dissociation to ensure faithful DNA replication. Mol Cell 44:85–96
Galbraith DW, Harkins KR, Maddox JM, Ayres NM, Sharma DP, Firoozabady E (1983) Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220:4601:1049–1051
Golbik R, Lupas AN, Koretke KK, Baumeister W, Peters J (1999) The Janus face of the archaeal Cdc48/p97 homologue VAT: protein folding versus unfolding. Biol Chem 380:1049–1062
Guinto JB, Ritson GP, Taylor JP, Forman MS (2007) Valosin-containing protein and the pathogenesis of frontotemporal dementia associated with inclusion body myopathy. Acta Neuropathol 114:55–61
Guo T, Chen K, Dong NQ et al (2018) Grain size and number1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30:871–888
Hartwell LH, Weinert TA (1989) Checkpoints: controls that ensure the order of cell cycle events. Science 246:629–634
Haubenberger D, Bittner RE, Rauch-Shorny S et al (2005) Inclusion body myopathy and Paget disease is linked to a novel mutation in the VCP gene. Neurology 65:1304–1305
Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protoc 3:824–834
Huang QN, Shi YF, Zhang XB, Song LX, Feng BH, Wang HM, Xu X, Li XH, Guo D, Wu JL (2016) Single base substitution in OsCDC48 is responsible for premature senescence and death phenotype in rice. J Integr Plant Biol 58:12–28
Imamura S, Ojima N, Yamashita M (2003) Cold-inducible expression of the cell division cycle gene CDC48 and its promotion of cell proliferation during cold acclimation in zebrafish cells. Febs Lett 549:14–20
Jentsch S, Rumpf S (2007) Cdc48 (p97): a ‘molecular gearbox’ in the ubiquitin pathway? Trends Biochem Sci 32:6–11
Kanehisa M, Araki M, Goto S et al (2008) KEGG for linking genomes to life and the environment. Nucleic Acids Res 36:D480–D484
Koller KJ, Brownstein MJ (1987) Use of a cDNA clone to identify a supposed precursor protein containing valosin. Nature 325:542–545
Kuhlbrodt K, Janiesch PC, Kevei E, Segref A, Barikbin R, Hoppe T (2011) The Machado-Joseph disease deubiquitylase ATX-3 couples longevity and proteostasis. Nat Cell Biol 13:273–455
Lamb JR, Fu V, Wirtz E, Bangs JD (2001) Functional analysis of the trypanosomal AAA protein TbVCP with trans-dominant ATP hydrolysis mutants. J Biol Chem 276:21512–21520
Leng YJ, Yang Y, Ren DY et al (2017) A Rice PECTATE LYASE-LIKE gene is required for plant growth and leaf senescence. Plant Physiol 174:1151–1166
Madeo F, Frohlich E, Frohlich KU (1997) A yeast mutant showing diagnostic markers of early and late apoptosis. J Cell Biol 139:729–734
Meyer H, Bug M, Bremer S (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat Cell Biol 14:117–123
Moir D, Stewart SE, Osmond BC, Botstein D (1982) Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics 100:547–563
Moradi F, Ismail AM (2007) Responses of photosynthesis, chlorophyll fluorescence and ROS-Scavenging systems to salt stress during seedling and reproductive stages in rice. Ann Bot Lond 99:1161–1173
Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B (2008) Mapping and quantifying mammalian transcriptomes by RNA-SEq. Nat Methods 5:621–628
Mouysset J, Deichsel A, Moser S, Hoege C, Hyman AA, Gartner A, Hoppe T (2008) Cell cycle progression requires the CDC-48(UFD-1/NPL-4) complex for efficient DNA replication. Proc Natl Acad Sci USA 105:12879–12884
Niwa H, Ewens CA, Tsang C, Yeung HO, Zhang XD, Freemont PS (2012) The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. J Biol Chem 287:8561–8570
Paro S, Rancour DM, Bednarek SY (2007) Protein domain-domain interactions and requirements for the negative regulation of Arabidopsis CDC48/p97 by the plant ubiquitin regulatory X (UBX) domain-containing protein, PUX1. J Biol Chem 282:5217–5224
Peters JM, Walsh MJ, Franke WW (1990) An abundant and ubiquitous homo-oligomeric ringshaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. EMBO J 9:1757–1767
Rockel B, Jakana J, Chiu W, Baumeister W (2002) Electron cryo-microscopy of VAT, the archaeal p97/CDC48 homologue from Thermoplasma acidophilum. J Mol Biol 317:673–681
Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM (2002) Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nat Struct Biol 9:950–957
Rubin D, Glickman M, Larsen C, Dhruvakumar S, Finley D (1998) Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J 17:4909–4919
Schmittgen TD, Livak KJ (2008) Analyzing real-time PCR data by the comparative C-T method. Nat Protoc 3:1101–1108
Shimizu S, Mori H (1998) Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant Cell Physiol 39:255–262
Shirogane T, Fukada T, Muller JMM, Shima DT, Hibi M, Hirano T (1999) Synergistic roles for Pim-1 and c-Myc in STAT3-mediated cell cycle progression and antiapoptosis. Immunity 11:709–719
Sugimoto M, Yamaguchi Y, Nakamura K, Tatsumi Y, Sano H (2004) A hypersensitive response-induced ATPase associated with various cellular activities (AAA) protein from tobacco plants. Plant Mol Biol 56:973–985
Thordal-Christensen H, Zhang ZG, Wei YD, Collinge DB (1997) Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J 11:1187–1194
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-SEq. Bioinformatics 25:1105–1111
Turner J, Hingorani M, Kelman Z, O’Donell M (1999) The internal workings of a DNA polymerase clamp-loading machine. EMBO J 18:771–783
Waadt R, Kudla J (2008) In Planta visualization of protein interactions using Bimolecular Fluorescence Complementation (BiFC). CSH Protoc 2008: pdb prot4995
Wang YH, Li JY (2011) Branching in rice. Curr Opin Plant Biol 14:94–99
Wang Q, Song CC, Li CCH (2003a) Hexamerization of p97-VCP is promoted by ATP binding to the D1 domain and required for ATPase and biological activities. Biochem Bioph Res Co 300:253–260
Wang Q, Song CC, Yang XY, Li CCH (2003b) D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. J Biol Chem 278:32784–32793
Wang Q, Song CC, Li CCH (2004) Molecular perspectives on p97-VCP: progress in understanding its structure and diverse biological functions. J Struct Biol 146:44–57
Wang C, Shen L, Fu YP, Yan CJ, Wang KJ (2015) A Simple CRISPR/Cas9 system for multiplex genome editing in rice. J Genet Genomics 42:703–706
Wellburn (1994) The spectral determination of chlorophyll a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J Plant Physiol Biochem 144:307–313
Wolf DH, Stolz A (2012) The Cdc48 machine in endoplasmic reticulum associated protein degradation. Mol Cell Res 1823:117–124
Xia D, Tang WK, Ye YH (2016) Structure and function of the AAA + ATPase p97/Cdc48p. Gene 583:64–77
Xu C, Wang YH, Yu YC et al (2012) Degradation of MONOCULM 1 by APC/C-TAD1 regulates rice tillering. Nat Commun. http:./doi.org/75010.1038/ncomms1743
Ye Y (2006) Diverse functions with a common regulator: Ubiquitin takes command of an AAA ATPase. J Struct Biol 156(2006):29–40
Yin ZC, Chen J, Zeng LR, Goh ML, Leung H, Khush GS, Wang GL (2000) Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol Plant Microbe Interact 13:869–876
Zhang XD, Shaw A, Beats PA et al (2000) Structure of the AAA ATPase p97. Mol Cell 6:1473–1484
Zhang Y, Su JB, Duan S et al (2011) A highly efficient rice green tissue protoplast system for transient gene expression and studying light/chloroplast-related processes. Plant Methods. https://doi.org/10.1186/1746-4811-7-30
Zhou F, Lin Q, Zhu L et al (2013) D14-SCF(D3)-dependent degradation of D53 regulates strigolactone signalling. Nature 504:406–410
Acknowledgements
This work was supported by the National Science Foundation of China (31701407), the Ministry of Science and Technology of China (2016YFD0101104). We thank Ms. Wenfang Zhao at the Institute of Plant Physiology and Ecology, SIBS, CAS, for technical support of flow cytometric analysis.
Author information
Authors and Affiliations
Contributions
QH and JW conceived and designed the research; LS, BZ, YS and XX performed the experiments; LS, QH, YH, and GS carried out the data analysis; LS, QH, YH, GS and JW wrote and revised the manuscript. All authors read and approve the final manuscript.
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Shi, L., Zhang, Xb., Shi, Yf. et al. OsCDC48/48E complex is required for plant survival in rice (Oryza sativa L.). Plant Mol Biol 100, 163–179 (2019). https://doi.org/10.1007/s11103-019-00851-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00851-9