Abstract
Key message
The novel AP2/ERF transcription factor SmERF128 positively regulates diterpenoid tanshinone biosynthesis by activating the expression of SmCPS1, SmKSL1, and SmCYP76AH1 in Salvia miltiorrhiza.
Abstract
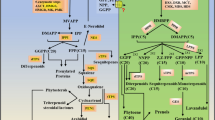
Certain members of the APETALA2/ethylene-responsive factor (AP2/ERF) family regulate plant secondary metabolism. Although it is clearly documented that AP2/ERF transcription factors (TFs) are involved in sesquiterpenoid biosynthesis, the regulation of diterpenoid biosynthesis by AP2/ERF TFs remains elusive. Here, we report that the novel AP2/ERF TF SmERF128 positively regulates diterpenoid tanshinone biosynthesis in Salvia miltiorrhiza. Overexpression of SmERF128 increased the expression levels of copalyl diphosphate synthase 1 (SmCPS1), kaurene synthase-like 1 (SmKSL1) and cytochrome P450 monooxygenase 76AH1 (SmCYP76AH1), whereas their expression levels were decreased when SmERF128 was silenced. Accordingly, the content of tanshinone was reduced in SmERF128 RNA interference (RNAi) hairy roots and dramatically increased in SmERF128 overexpression hairy roots, as demonstrated through Ultra Performance Liquid Chromatography (UPLC) and Liquid Chromatography-Tandem Mass Spectrometry (LC-MS/MS) analysis. Furthermore, SmERF128 activated the expression of SmCPS1, SmKSL1, and SmCYP76AH1 by binding to the GCC box, and to the CRTDREHVCBF2 (CBF2) and RAV1AAT (RAA) motifs within their promoters during in vivo and in vitro assays. Our findings not only reveal the molecular basis of how the AP2/ERF transcription factor SmERF128 regulates diterpenoid biosynthesis, but also provide useful information for improving tanshinone production through genetic engineering.






Similar content being viewed by others
Abbreviations
- AP2/ERF:
-
APETALA2/ethylene-responsive factor
- CMK :
-
4-(Cytidine 5′-diphospho)-2-C-methyl-D-erythritol kinase
- CPS1 :
-
Copalyl diphosphate synthase 1
- CYP76AH1 :
-
Cytochrome P450 monooxygenase 76AH1
- DMAPP:
-
Dimethylallyl diphosphate
- DXR :
-
1-Deoxy-D-xylulose-5-phosphate reductoisomerase
- DXS2 :
-
1-Deoxy-D-xylulose-5-phosphate synthase 2
- GFP:
-
Green fluorescent protein
- GGPP:
-
Geranylgeranyl diphosphate
- GGPPS :
-
Geranylgeranyl diphosphate synthase
- GUS:
-
β-Glucuronidase
- HDR1 :
-
4-Hydroxy-3-methylbut-2-enyl diphosphate reductase 1
- HDS :
-
4-Hydroxy-3-methylbut-2-enyl diphosphate synthase
- IPP:
-
Isopentenyl diphosphate
- IPTG:
-
Isopropyl β-D-thiogalactoside
- KSL1 :
-
Kaurene synthase-like 1
- MCT :
-
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase
- MDS :
-
2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase
- NADPH:
-
Nicotinamide adenine dinucleotide phosphate
- OX:
-
Overexpression
- RNAi:
-
RNA interference
- TIA:
-
Terpenoid indole alkaloid
- TF:
-
Transcription factor
References
Adachi H, Nakano T, Miyagawa N et al (2015) WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. Plant Cell 27:2645–2663
Chang CC, Chu CF, Wang CN et al (2014) The anti-atherosclerotic effect of tanshinone IIA is associated with the inhibition of TNF-α-induced VCAM-1, ICAM-1 and CX3CL1 expression. Phytomedicine 21:207–216
Chen S, Song J, Sun C, Xu J, Zhu Y, Verpoorte R, Fan T (2015) Herbal genomics: examining the biology of traditional medicines. Science 347:S27–S29
Cheng TO (2006) Danshen: a popular Chinese cardiac herbal drug. J Am Coll Cardiol 47:1498
Cui G, Duan L, Jin B et al (2015) Functional divergence of diterpene syntheses in the medicinal plant Salvia miltiorrhiza. Plant Physiol 169:1607–1618
Dai Y, Qin Q, Dai D, Kong L, Li W, Zha X, Jin Y, Tang K (2009) Isolation and characterization of a novel cDNA encoding methyl jasmonate-responsive transcription factor TcAP2 from Taxus cuspidata. Biotechnol Lett 31:1801–1809
De Boer K, Tilleman S, Pauwels L et al (2011) APETALA2/ETHYLENE RESPONSE FACTOR and basic helix-loop-helix tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J 66:1053–1065
Ding K, Pei T, Bai Z, Jia Y, Ma P, Liang Z (2017) SmMYB36, a Novel R2R3-MYB transcription factor, enhances tanshinone accumulation and decreases phenolic acid content in Salvia miltiorrhiza hairy roots. Sci Rep 7:5104
Dong Y, Morris-Natschke SL, Lee KH (2011) Biosynthesis, total syntheses, and antitumor activity of tanshinones and their analogs as potential therapeutic agents. Nat Prod Rep 28:529–542
IdrovoEspin FM, Peraza-Echeverria S, Fuentes G, Santamaria JM (2012) In silico cloning and characterization of the TGA (TGACG MOTIF-BINDING FACTOR) transcription factors subfamily in Carica papaya. Plant Physiol Biochem 54:113–122
Ji A, Luo H, Xu Z et al (2016) Genome-wide identification of the AP2/ERF gene family involved in active constituent biosynthesis in Salvia miltiorrhiza. Plant Genome 9(2):1–11
Liu X, Guo CY, Ma XJ et al (2015) Anti-inflammatory effects of tanshinone IIA on atherosclerostic vessels of ovariectomized ApoE mice are mediated by estrogen receptor activation and through the ERK signaling pathway. Cell Physiol Biochem 35:1744–1755
Lu X, Zhang L, Zhang F et al (2013) AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol 198:1191–1202
Ma Y, Yuan L, Wu B, Li X, Chen S, Lu S (2012) Genome-wide identification and characterization of novel genes involved in terpenoid biosynthesis in Salvia miltiorrhiza. J Exp Bot 63:2809–2823
Naseri G, Balazadeh S, Machens F, Kamranfar I, Messerschmidt K, Mueller-Roeber B (2017) Plant-derived transcription factors for orthologous regulation of gene expression in the yeast saccharomyces cerevisiae. Acs Synth Biol 6:1742–1756
Paul P, Singh SK, Patra B, Sui X, Pattanaik S, Yuan L (2017) A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol 213:1107–1123
Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290:998–1009
Sears MT, Zhang H, Rushton PJ et al (2014) NtERF32: a non-NIC2 locus AP2/ERF transcription factor required in jasmonate-inducible nicotine biosynthesis in tobacco. Plant Mol Biol 84:49–66
Shoji T, Hashimoto T (2011) Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol 52:1117–1130
Shoji T, Kajikawa M, Hashimoto T (2010) Clustered transcription factor genes regulate nicotine biosynthesis in tobacco. Plant Cell 22:3390–3409
Tan H, Xiao L, Gao S et al (2015) TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol Plant 8:1396–1411
Tao S, Zheng Y, Lau A et al (2013) Tanshinone I activates the Nrf2-dependent antioxidant response and protects against as (III)-induced lung inflammation in vitro and in vivo. Antioxid Redox Signal 19:1647–1661
Van der Fits L, Memelink J (2000) ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289:295–297
Van der Fits L, Memelink J (2001) The jasmonate-inducible AP2/ERF domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J 25:43–53
Wang X, Morris-Natschke SL, Lee KH (2007) New developments in the chemistry and biology of the bioactive constituents of Tanshen. Med Res Rev 27:133–148
Xin T, Zhang Y, Pu X, Gao R, Xu Z, Song J (2018) Trends in herbgenomics. Sci China Life Sci. https://doi.org/10.1007/s11427-018-9352-7
Xu Z, Song J (2017) The 2-oxoglutarate-dependent dioxygenase superfamily participates in tanshinone production in Salvia miltiorrhiza. J Exp Bot 68:2299–2308
Xu Y, Wang Y, Long Q et al (2014) Overexpression of OsZHD1, a zinc finger homeodomain class homeobox transcription factor, induces abaxially curled and drooping leaf in rice. Planta 239:803–816
Xu Z, Peters RJ, Weirather J et al (2015) Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J 82:951–961
Xu H, Song J, Luo H et al (2016a) Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol Plant 9:949–952
Xu Z, Ji A, Zhang X, Song J, Chen S (2016b) Biosynthesis and regulation of active compounds in medicinal model plant Salvia miltiorrhiza. Chin Herb Med 8:3–11
Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2:1565–1572
Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ, Chen XY (2012) The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant 5:353–365
Zhang H, Hedhili S, Montiel G, Zhang Y, Chatel G, Pre M, Gantet P, Memelink J (2011) The basichelix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J 67:61–71
Zhang HB, Bokowiec MT, Rushton PJ, Han SC, Timko MP (2012a) Tobacco transcription factors NtMYC2a and NtMYC2b form nuclear complexes with the NtJAZ1 repressor and regulate multiple jasmonate-inducible steps in nicotine biosynthesis. Mol Plant 5:73–84
Zhang Y, Jiang P, Ye M et al (2012b) Tanshinones: sources, pharmacokinetics and anti-cancer activities. In J Mol Sci 13:13621–13666
Zhang X, Luo H, Xu Z, Zhu Y, Ji A, Song J, Chen S (2015) Genome-wide characterisation and analysis of bHLH transcription factors related to tanshinone biosynthesis in Salvia miltiorrhiza. Sci Rep 5:11244
Zhang J, Zhou L, Zheng X, Zhang J, Yang L, Tan R, Zhao S (2017) Overexpression of SmMYB9b enhances tanshinone concentration in Salvia miltiorrhiza hairy roots. Plant Cell Rep 36:1297–1309
Zhang Y, Xu Z, Ji A, Luo H, Song J (2018) Genomic survey of bZIP transcription factor genes related to tanshinone biosynthesis in Salvia miltiorrhiza. Acta Pharm Sin B 8:295–305
Zheng J, Lang Y, Zhang Q et al (2015) Structure of human MDM2 complexed with RPL11 reveals the molecular basis of p53 activation. Gene Dev 29:1524–1534
Zhou Y, Sun W, Chen J et al (2016) SmMYC2a and SmMYC2b played similar but irreplaceable roles in regulating the biosynthesis of tanshinones and phenolic acids in Salvia miltiorrhiza. Sci Rep 6:22852
Zhu D, Wu Z, Cao G et al (2014) Translucent green, an ERF family transcription factor, controls water balance in Arabidopsis by activating the expression of aquaporin genes. Mol Plant 7:601–615
Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81573398), CAMS Innovation Fund for Medical Sciences (CIFMS) (2017-I2M-1-009) and Foundation of Educational Department of Guangdong Province (E1-KFD015181K31).
Author information
Authors and Affiliations
Contributions
JS designed the study. YZ and AJ performed experiments. YZ, AJ, ZX and HL analyzed the data. YZ, AJ, JS, and ZX wrote the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare that they have no conflict of interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhang, Y., Ji, A., Xu, Z. et al. The AP2/ERF transcription factor SmERF128 positively regulates diterpenoid biosynthesis in Salvia miltiorrhiza. Plant Mol Biol 100, 83–93 (2019). https://doi.org/10.1007/s11103-019-00845-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-019-00845-7