Abstract
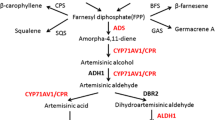
The artemisinic aldehyde double bond reductase (DBR2) plays an important role in the biosynthesis of the antimalarial artemisinin in Artemisia annua. Artemisinic aldehyde is reduced into dihydroartemisinic aldehyde by DBR2. Artemisinic aldehyde can also be oxidized by amorpha-4,11-diene 12-hydroxylase and/or aldehyde dehydrogenase 1 to artemisinic acid, a precursor of arteannuin B. In order to better understand the effects of DBR2 expression on the flow of artemisinic aldehyde into either artemisinin or arteannuin B, we determined the content of dihydroartemisinic aldehyde, artemisinin, artemisinic acid and arteannuin B content of A. annua varieties sorted into two chemotypes. The high artemisinin producers (HAPs), which includes the ‘2/39’, ‘Chongqing’ and ‘Anamed’ varieties, produce more artemisinin than arteannuin B; the low artemisinin producers (LAPs), which include the ‘Meise’, ‘Iran#8’, ‘Iran#14’, ‘Iran#24’ and ‘Iran#47’ varieties, produce more arteannuin B than artemisinin. Quantitative PCR showed that the relative expression of DBR2 was significantly higher in the HAP varieties. We cloned and sequenced the promoter of the DBR2 gene from varieties of both the LAP and the HAP groups. There were deletions/insertions in the region just upstream of the ATG start codon in the LAP varities, which might be the reason for the different promoter activities of the HAP and LAP varieties. The relevance of promoter variation, DBR2 expression levels and artemisinin biosynthesis capabilities are discussed and a selection method for HAP varieties with a DNA marker is suggested. Furthermore, putative cis-acting regulatory elements differ between the HAP and LAP varieties.







Similar content being viewed by others
Abbreviations
- AA:
-
Artemisinic acid
- AAld:
-
Artemisinic aldehyde
- AB:
-
Arteannuin B
- ABA:
-
Abscisic acid
- ADH1:
-
Alcohol dehydrogenase 1
- ADS:
-
Amorpha-4,11-diene synthase
- bHLH:
-
Basic/helix-loop-helix
- ALDH1:
-
Aldehyde dehydrogenase 1
- AP2:
-
APETALA2
- ART:
-
Artemisinin
- CaMV:
-
Cauliflower mosaic virus
- CPR:
-
Cytochrome P450 reductase
- CYP71AV1:
-
Amorpha-4,11-diene 12-hydroxylase
- DBR2:
-
Artemisinic aldehyde Δ11(13) reductase
- ERF:
-
Ethylene response factor
- DHAA:
-
Dihydroartemisinic acid
- DHAAld:
-
Dihydroartemisinic aldehyde
- FAR:
-
β-Farnesene
- FDS:
-
Farnesyl diphosphate synthase
- GA:
-
Gibberellinic acid
- GSP:
-
Gene specific primer
- GST:
-
Glandular secretory trichome
- HAP:
-
High artemisinin producer
- HMGR:
-
3-Hydroxy-3-methyl-glutaryl-CoA reductase
- IDI:
-
Isopentenyldiphosphate isomerase
- JA:
-
Jasmonate
- LAP:
-
Low artemisinin producer
- MeJA:
-
Methyl jasmonate
- MYB:
-
MYB transcription factor
- OPR:
-
12-Oxophytodienoate reductase
- SA:
-
Salicylic acid
- TSS:
-
Transcription start site
- WRKY:
-
WRKY transcription factor
References
Alam P, Abdin MZ (2011) Over-expression of HMG-CoA reductase and amorpha-4,11-diene synthase genes in Artemisia annua L. and its influence on artemisinin. Plant Cell Rep 30:1919–1928
Baldi A, Dixit VK (2008) Yield enhancement strategies for artemisinin production by suspension cultures of Artemisia annua. Biores Technol 99:4609–4614
Banyai W, Mii M, Supaibulwatana K (2011) Enhancement of artemisinin content and biomass in Artemisia annua by exogenous GA3 treatment. Plant Growth Reg 63:45–54
Bertea CM, Freije JR, van der Woude H, Verstappen FW, Perk L, Marquez V, De Kraker JW, Posthumus MA, Jansen BJ, de Groot A, Franssen MC, Bouwmeester HJ (2005) Identification of intermediates and enzymes involved in the early steps of artemisinin biosynthesis in Artemisia annua. Planta Med 71:40–47
Boter M, Ruiz-Rivero O, Abdeen A, Prat S (2004) Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev 18:1577–1591
Brown GD (2010) The biosynthesis of artemisinin (Qinghaosu) and the phytochemistry of Artemisia annua L. (Qinghao). Molecules 15:7603–7698
Brown GD, Sy L-K (2004) In vivo transformations of dihydroartemisinic acid in Artemisia annua plants. Tetrahedron 60:1139–1159
Brown GD, Sy L-K (2006) In vivo transformations of artemisinic acid in Artemisia annua plants. Tetrahedron 60:1139–1159
Caretto S, Quarta A, Durante M, Nisi R, De Paolis A, Blando F, Mita G (2011) Methyl jasmonate and miconazole differently affect arteminisin production and gene expression in Artemisia annua suspension cultures. Plant Biol 13:51–58
Covello PS (2008) Making artemisinin. Phytochemistry 69:2881–2885
Covello PS, Teoh KH, Polichuk DR, Reed DW, Nowak G (2007) Functional genomics and the biosynthesis of artemisinin. Phytochemistry 68:1864–1871
Duke MV, Paul RN, Elsohly HN, Sturtz G, Duke SO (1994) Localization of artemisinin and artemisitene in foliar tissues of glanded and glandless biotypes of Artemisia annua L. Int J Plant Sci 155:365–372
Guom X-X, Yang X-Q, Yang R-Y, Zeng Q-P (2010) Salicylic acid and methyl jasmonate but not Rose Bengal enhance artemisinin production through invoking burst of endogenous singlet oxygen. Plant Sci 178:390–397
Han J, Wang H, Lundgren A, Brodelius PE (2014) Effects of overexpression of AaWRKY1 on artemisinin biosynthesis in transgenic Artemisia annua plants. Phytochemistry 102:89–96
Ji J Xiao, Shen Y, Ma D, Li Z, Pu G, Li X, Huang L, Liu B, Ye H, Wang H (2014) Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol 55:1592–1604
Jiang W, Lu X, Qiu B, Zhang F, Shen Q, Lv Z, Fu X, Yan T, Gao E, Zhu M, Chen L, Zhang L, Wang G, Sun X, Tang K (2014) Molecular cloning and characterization of a trichome-specific promoter of artemisinic aldehyde Δ11(13) reductase (DBR2) in Artemisia annua. Plant Mol Biol Rep 32:82–91
Jing F, Zhang L, Li M, Tang Y, Wang Y, Wang Y, Wang Q, Pan Q, Wang G, Tang K (2009) Abscisic acid (ABA) treatment increases artemisinin content in Artemisia annua by enhancing the expression of genes in artemisinin biosynthetic pathway. Biologia 64:319–323
Kim J, Kim HY (2006) Molecular characterization of a bHLH transcription factor involved in Arabidopsis abscisic acid-mediated response. Biochim Biophys Acta 1759:191–194
Lei C, Ma D, Pu G, Qiu X, Du Z, Wang H, Li G, Ye H, Liu B (2011) Foliar application of chitosan activates artemisinin biosynthesis in Artemisia annua L. Ind Crops Prod 33:176–182
Lescot M, Déhais P, Moreau Y, De Moor B, Rouzé P, Rombauts S (2002) PlantCARE: a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30:325–327
Lu X, Zhang L, Zhang F, Jiang W, Shen Q, Zhang L, Lv Z, Wang G, Tang K (2013) AaORA, a trichome-specific AP2/ERF transcription factor of Artemisia annua, is a positive regulator in the artemisinin biosynthetic pathway and in disease resistance to Botrytis cinerea. New Phytol 198:1191–1202
Ma D, Pu G, Lei C, Ma L, Wang H, Guo Y, Chen J, Du Z, Wang H, Li G, Ye H, Liu B (2009a) Isolation and characterization of AaWRKY1, an Artemisia annua transcription factor that regulates the amorpha-4,11-diene synthase gene, a key gene of artemisinin biosynthesis. Plant Cell Physiol 50:2146–2161
Ma C, Wang H, Lu X, Wang H, Xu G, Liu B (2009b) Terpenoid metabolic profiling analysis of transgenic Artemisia annua L. by comprehensive two-dimensional gas chromatography time-of-flight mass spectrometry. Metabolomics 5:497–506
Maes L, van Nieuwerburgh FCW, Zhang Y, Reed DW, Pollier J, Vande Casteele SRF, Inzé D, Covello PS, Deforce DLD, Goossens A (2011) Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol 189:176–189
McGarvey DJ, Croteau R (1995) Terpenoid metabolism. Plant Cell 7:1015–1026
Miyamoto K, Shimizu T, Lin FQ, Sainsbury F, Thuenemann E, Lomonossoff G, Nojiri H, Yamane H, Okada K (2012) Identification of an E-box motif responsible for the expression of jasmonic acid-induced chitinase gene OsChia4a in rice. J Plant Physiol 169:621–627
Narusaka Y, Nakashima K, Shinwari ZK, Sakuma Y, Furihata T et al (2003) Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J 34:137–148
Olofsson L, Engström A, Lundgren A, Brodelius EP (2011) Relative epression of genes of terpene metabolism in different tissues of Artemisia annua L. BMC Plant Biol 11:45
Olofsson L, Lundgren A, Brodelius PE (2012) Trichome isolation with and without fixation using laser microdissection and pressure catapulting followed by RNA amplification: expression of genes of terpene metabolism in apical and sub-apical trichome cells of Artemisia annua L. Plant Sci 183:9–13
Paddon CJ, Westfall PJ, Pitera DJ, Benjamin K, Fisher K, McPhee D, Leavell MD, Tai A, Main D, Eng D, Polichuk DR, Teoh KH, Reed DW, Treynor T, Lenihan J, Jiang H, Fleck M, Bajad S, Dang G, Dengrove D, Diola D, Ellens KW, Fickes S, Galazzo J, Gaucher SP, Geistlinger T, Henry R, Hepp M, Horning T, Iqbal T, Jiang H, Kizer L, Lieu B, Melis D, Moss N, Regentin R, Secrest S, Tsuruta H, Vazquez R, Westblade LF, Xu L, Yu M, Zhang Y, Zhao L, Lievense J, Covello PS, Keasling JD, Reiling KK, Renninger NS, Newman JD (2013) High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496:528–532
Pu GB, Ma DM, Chen JL, Ma LQ, Wang H, Li GF, Ye HC, Liu BY (2009) Salicylic acid activates artemisinin biosynthesis in Artemisia annua L. Plant Cell Rep 28:1127–1135
Putalun W, Luealon W, De-Eknamkul W, Tanaka H, Shoyama Y (2007) Improvement of artemisinin production by chitosan in hairy root cultures of Artemisia annua L. Biotechnol Lett 29:1143–1146
RBM/UNITAID/WHO (2011) Artemisinin Conference, Hanoi, Viet Nam, November 2–3, 2011. http://www.mmv.org/sites/default/files/uploads/docs/events/2011/2011_Artemisinin_Conference_Report.pdf
Reyes JC, Muro-Pastor MI, Florencio FJ (2004) The GATA family of transcription factors in Arabidopsis and rice. Plant Physiol 134:1718–1732
Rushton PJ, Somssich IE, Ringler P, Shen QJ (2010) WRKY transcription factors. Trends Plant Sci 15:247–258
Sarvestani R, Peyghambary SA, Abbasi A (2014) Isolation and characterization of DBR2 gene promoter from Iranian Artemisia annua. J Agr Sci Tech 16:191–202
Shen QX, Ho THD (1995) Functional dissection of an abscisic-acid (ABA)-inducible gene reveals 2 independent ABA-responsive complexes each containing a G-box and a novel cis-acting element. Plant Cell 7:295–307
Shen QX, Zhang PN, Ho THD (1996) Modular nature of abscisic acid (ABA) response complexes: composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell 8:1107–1119
Sipahimalani AT, Fulzele DP, Heble MR (1991) Rapid method for the detection and determination of artemisinin by gas chromatography. J Chromatogr A 538:452–455
Solano R, Nieto C, Avila J, Cañas L, Diaz I, Paz-Ares J (1995) Dual DNA binding specificity of a petal epidermis-specific MYB transcription factor (MYB. Ph3) from Petunia hybrid. EMBO J 14:1773–1784
Teoh KH, Polichuk DR, Reed DW, Nowak G, Covello PS (2006) Specific cDNAs reveal CYP71AV1, a cytochrome P450 with a key role in the biosynthesis of the antimalarial sesquiterpene lactone artemisinin. FEBS Lett 580:1411–1416
Teoh KH, Polichuk DR, Reed DW, Covello PS (2009) Molecular cloning of an aldehyde dehydrogenase implicated in artemisinin biosynthesis in Artemisia annua. Botany 87:635–642
Ting H-M, Wang B, Ryden A-M, Woittiez L, van Herpen T, Verstappen FWA, Ruyter-Spira C, Jules Beekwilder J, Bouwmeester HJ, van der Krol A (2013) The metabolite chemotype of Nicotiana benthamiana transiently expressing artemisinin biosynthetic pathway genes is a function of CYP71AV1 type and relative gene dosage. New Phytol 199:352–366
Urao T, Yamaguchi-Shinozaki K, Urao S, Shinozaki K (1993) An Arabidopsis myb homolog is induced by dehydration stress and its gene product binds to the conserved MYB recognition sequence. Plant Cell 5:1529–1539
Wallaart TE, Bouwmeester HJ, Hille J, Poppinga L, Maijers NC (2000) Amorpha-4,11-diene synthase: cloning and functional expression of a key enzyme in the biosynthetic pathway of the novel antimalarial drug artemisinin. Planta 212:460–465
Wang H, Olofsson L, Lundgren A, Brodelius PE (2011) Trichome-specific expression of amorpha-4,11-diene synthase, a key enzyme of artemisinin biosynthesis in Artemisia annua L., as reported by a promoter-GUS fusion. Am J Plant Sci 2:619–628
Wang H, Han J, Kanagarajan S, Lundgren A, Brodelius PE (2013) Trichome-specific expression of the amorpha-4,11-diene 12-hydroxylase (cyp71av1) gene, encoding a key enzyme of artemisinin biosynthesis in Artemisia annua, as reported by a promoter-GUS fusion. Plant Mol Biol 81:119–138
White NJ (1997) Assessment of the pharmacodynamic properties of antimalarial drugs in vivo. Antimicrob Agents Chemother 47:1413–1422
Woerdenbag HJ, Pras N, Bos R, Visser JF, Hendriks H, Malingré TM (1991) Analysis of artemisinin and related sesquiterpenoids from Artemisia annua L. by combined gas chromatography/mass spectrometry. Phytochem Anal 2:215–219
World Health Organization (2014) World Malaria Report 2014 http://www.who.int/malaria/publications/world_malaria_report_2014/en/
Wu W, Yuan M, Zhang Q, Zhu YM, Yong L et al (2011) Chemotype-dependent metabolic response to methyl jasmonate elicitation in Artemisia annua. Planta Med 77:1048–1053
Xu Y-H, Wang J-W, Wang S, Wang J-Y, Chen X-Y (2004) Characterization of GaWRKY1, a cotton transcription factor that regulates the sesquiterpene synthase gene (+)-γ-cadinene synthase-A. Plant Physiol 135:507–515
Yang TB, Poovaiah BW (2001) A calmodulin-binding/CGCG box DNA- binding protein family involved in multiple signaling pathways in plants. J Biol Chem 277:45049–45058
Yu ZX, Li JX, Yang CQ, Hu WL, Wang LJ et al (2012) The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol Plant 5:353–365
Yuan Y, Liu W, Zhang Q, Xiang L, Liu X, Chen M, Lin Z, Wang Q, Liao Z (2014) Overexpression of artemisinic aldehyde Δ11(13) reductase gene-enhanced artemisinin and its relative metabolite biosynthesis in transgenic Artemisia annua L. Biotechnol Appl Biochem. doi:10.1002/bab.1234
Zare Mehrjerdi M, Bihamta M-R, Omidi M, Naghavi M-R, Soltanloo H, Ranjbar M (2013) Effects of exogenous methyl jasmonate and 2-isopentenyladenine on artemisinin production and gene expression in Artemisia annua. Turk J Bot 37:499–505
Zhang YS, Ye HC, Liu BY, Wangand H, Li GF (2005) Exogenous GA3 and flowering induce the conversion of artemisinic acid to artemisinin in Artemisia annua plants Russ. J Plant Physiol 52:58–62
Zhang Y, Teoh KH, Reed DW, Maes L, Goossens A, Olson DJH, Ross ARS, Covello PS (2008) The molecular cloning of artemisinic aldehyde Δ11(13) reductase and its role in glandular trichome-dependent biosynthesis of artemisinin in Artemisia annua. J Biol Chem 283:21501–21508
Zhang L, Jing F, Li F, Li M, Wang Y, Wang G, Sun X, Tang K (2009) Development of transgenic Artemisia annua (Chinese wormwood) plants with an enhanced content of artemisinin, an effective anti-malarial drug, by hairpin-RNA-mediated gene silencing. Biotechnol Appl Biochem 52:199–207
Acknowledgments
We would like to thank Professor Kexuan Tang of Shanghai Jiao Tong University for technical advice and assistance. We also thank Tehran University and Ghent University for providing A. annua seeds. This work was supported by the Faculty of Life and Health Sciences, Linnaeus University.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
11103_2015_284_MOESM1_ESM.pdf
Figure 1S. GC–MS analysis of artemisinin and arteannuin B and their biosynthetic precursors. A: Artemisinin standard showing the thermal breakdown into 3 products; B: Separation of precursors of artemisinin and arteannuin B biosynthesis; C: Separation of artemisinin and arteannuin B (PDF 241 kb)
11103_2015_284_MOESM3_ESM.pdf
Figure 3S. Nucleotide sequence of the DBR2 gene from the ‘Anamed’ variety of Artemisia annua. UPPERCASE: exons; lowercase: introns (PDF 37 kb)
11103_2015_284_MOESM5_ESM.pdf
Figure 5S. Alignment of the nucleotide sequences of the 3′-end of the DBR2 promoters amplified by PCR (variable region) (cf. Figure 7). Putative cis-acting regulatory elements are shown in different colours. → indicates that the putative cis-acting element is located to the leading strand; ← indicates that the putative cis-acting element is located to the lagging strand; ↔ indicates that the putative cis-acting element is located to both strands (PDF 76 kb)
11103_2015_284_MOESM6_ESM.pdf
Figure 6S. Alignment of cloned fragment and cDNA and EST sequences from the NBCI GenBank as indicated. The sequences carry the 3′-end of promoters and the 5′-end of the open reading frames. The ATG start codon is marked with *** (PDF 30 kb)
11103_2015_284_MOESM7_ESM.pdf
Figure 7S. Alignment of the conserved nucleotide sequences of the DBR2 promoters amplified by PCR (cf. Figure 5). Putative cis-acting regulatory elements are shown in different colours. → indicates that the putative cis-acting element is located to the leading strand; ← indicates that the putative cis-acting element is located to the lagging strand; ↔ indicates that the putative cis-acting element is located to both strands (PDF 81 kb)
Rights and permissions
About this article
Cite this article
Yang, K., Monafared, R.S., Wang, H. et al. The activity of the artemisinic aldehyde Δ11(13) reductase promoter is important for artemisinin yield in different chemotypes of Artemisia annua L.. Plant Mol Biol 88, 325–340 (2015). https://doi.org/10.1007/s11103-015-0284-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11103-015-0284-3