Abstract
Purpose
To describe phenotype-genotype data of Asian-Indian normosmic congenital hypogonadotropic hypogonadism (nCHH) from our centre and perform a systematic review of genetic studies using next-generation sequencing (NGS) in nCHH.
Methods
Sixty-eight nCHH probands from our center, and 370 nCHH probands from published studies were included. Per-patient genetic variants were analyzed as per ACMG guidelines. Molecular diagnosis was defined as presence of a pathogenic or likely pathogenic variant in a known CHH gene following zygosity status as per known mode of genetic inheritance.
Result
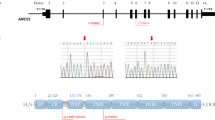
At our centre molecular diagnosis was observed in 35.3% of probands {GNRHR:16.2%, FGFR1:7.3%, KISS1R:4.4%, GNRH1:2.9%, TACR3:2.9%, CHD7:1.4%}. Molecular diagnosis was observed more often (44.7% vs 14.3%, p = 0.026) with severe than partial reproductive-phenotype. The study adds 12 novel variants and suggests GNRHR p.Thr32Ala variant may have a founder effect. In per-patient systematic review (including our cohort), the molecular diagnosis was reached in 23.2%, ranging from 3.5 to 46.7% at different centers. The affected genes were FGFR1:6.4%, GNRHR:4.3%, PROKR2:3.6%, TACR3:1.8%, CHD7:1.6%, KISS1R:1.4%, GNRH1:1.4% and others (PROK2, SOX3, SOX10, SOX11, IL17RD, IGSF10, TAC3, ANOS1, oligogenic): < 1% each. FGFR1 was the most commonly affected gene in most cohorts except Asia, whereas PROKR2 (in China and Japan) and GNRHR (in India) were the commonest.
Conclusion
(s): The global molecular diagnosis rate was 23.2% in nCHH cohorts whereas that in our cohort was 35% with a higher rate (44.7%) in those with severe reproductive-phenotype. The most commonly affected gene in nCHH patients was FGFR1 globally while it was PROKR2 in East Asia and GNRHR in India.

Similar content being viewed by others
References
Boehm U, Bouloux P-M, Dattani MT et al (2015) European Consensus Statement on congenital hypogonadotropic hypogonadism—pathogenesis, diagnosis and treatment. Nat Rev Endocrinol 11:547–564. https://doi.org/10.1038/nrendo.2015.112
Butz H, Nyírő G, Kurucz PA et al (2021) Molecular genetic diagnostics of hypogonadotropic hypogonadism: from panel design towards result interpretation in clinical practice. Hum Genet 140:113–134. https://doi.org/10.1007/s00439-020-02148-0
Maione L, Dwyer AA, Francou B et al (2018) GENETICS IN ENDOCRINOLOGY: genetic counseling for congenital hypogonadotropic hypogonadism and Kallmann syndrome: new challenges in the era of oligogenism and next-generation sequencing. Eur J Endocrinol 178:R55–R80. https://doi.org/10.1530/EJE-17-0749
Izumi Y, Suzuki E, Kanzaki S et al (2014) Genome-wide copy number analysis and systematic mutation screening in 58 patients with hypogonadotropic hypogonadism. Fertil Steril 102:1130-1136.e3. https://doi.org/10.1016/j.fertnstert.2014.06.017
Wang Y, Gong C, Qin M et al (2017) Clinical and genetic features of 64 young male paediatric patients with congenital hypogonadotropic hypogonadism. Clin Endocrinol 87:757–766. https://doi.org/10.1111/cen.13451
Cassatella D, Howard SR, Acierno JS et al (2018) Congenital hypogonadotropic hypogonadism and constitutional delay of growth and puberty have distinct genetic architectures. Eur J Endocrinol 178:377–388. https://doi.org/10.1530/EJE-17-0568
Zhou C, Niu Y, Xu H et al (2018) Mutation profiles and clinical characteristics of Chinese males with isolated hypogonadotropic hypogonadism. Fertil Steril 110:486-495.e5. https://doi.org/10.1016/j.fertnstert.2018.04.010
Kim JH, Seo GH, Kim G-H et al (2019) Targeted Gene Panel Sequencing for Molecular Diagnosis of Kallmann Syndrome and Normosmic Idiopathic Hypogonadotropic Hypogonadism. Exp Clin Endocrinol Diabetes 127:538–544. https://doi.org/10.1055/a-0681-6608
Amato LGL, Montenegro LR, Lerario AM et al (2019) New genetic findings in a large cohort of congenital hypogonadotropic hypogonadism. Eur J Endocrinol 181:103–119. https://doi.org/10.1530/EJE-18-0764
Stamou MI, Varnavas P, Plummer L et al (2019) Next-generation sequencing refines the genetic architecture of Greek GnRH-deficient patients. Endocr Connect 8:468–480. https://doi.org/10.1530/EC-19-0010
Cho C-Y, Tsai W-Y, Lee C-T et al (2021) Clinical and molecular features of idiopathic hypogonadotropic hypogonadism in Taiwan: a single center experience. J Formos Med Assoc. https://doi.org/10.1016/j.jfma.2021.03.010
Turkyilmaz A, Cayir A, Yarali O et al (2021) Clinical characteristics and molecular genetic analysis of a cohort with idiopathic congenital hypogonadism. J Pediatr Endocrinol Metab. https://doi.org/10.1515/jpem-2020-0590
Nair S, Jadhav S, Lila A et al (2016) Spectrum of phenotype and genotype of congenital isolated hypogonadotropic hypogonadism in Asian Indians. Clin Endocrinol 85:100–109. https://doi.org/10.1111/cen.13009
VarSome Citations (2020) In: VarSome. https://varsome.com/about/general/varsome-citations/. Accessed 15 May 2021
on behalf of the ACMG Laboratory Quality Assurance Committee, Richards S, Aziz N, et al (2015) Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med 17:405–423. https://doi.org/10.1038/gim.2015.30
Cangiano B, Swee DS, Quinton R, Bonomi M (2021) Genetics of congenital hypogonadotropic hypogonadism: peculiarities and phenotype of an oligogenic disease. Hum Genet 140:77–111. https://doi.org/10.1007/s00439-020-02147-1
Gürbüz F, Kotan LD, Mengen E et al (2012) Distribution of gene mutations associated with familial normosmic idiopathic hypogonadotropic hypogonadism. Jcrpe 4:121–126. https://doi.org/10.4274/Jcrpe.725
Acknowledgements
Clinical Exome Sequencing was performed at GenePath Diagnostics, Pune, India.
Funding
Ministry of Science and Technology, Department of Biotechnology, Government of India. (Grant -No.BT/PR8388/MED/12/620/2013).
Author information
Authors and Affiliations
Contributions
Contributors: NSS, ARL, and TRB conceived the idea and designed the study. VAP and NSS wrote the first draft of the report with input from ARL, SA, SJ, and TRB. AVE provided an expert review of the genetic results. VS, SJ, and SM accessed and verified the data, VAP and RS performed the statistical analysis which was verified by NSS and ARL. VAP, RS, ARL, NSS, TRB were involved in the management of patients and data collection. NSS, ARL, SM, SA, and TRB supervised the entire data collection and management and provided inputs in the revision of the draft. All the authors were involved in the critical revision of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All procedures involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the Helsinki declaration and its later amendments or comparable ethical standards. This study was approved by Institutional Ethics Committee III of Seth G S Medical College and KEM hospital (EC/159/2009). There was no role or influence of the funder in study design, collection, analysis, and interpretation of data, writing of the report, or submission of the report for publication.
Informed consent
Informed consent was obtained from all individual participants involved in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patil, V.A., Lila, A.R., Shah, N. et al. Regional genotypic variations in normosmic congenital hypogonadotropic hypogonadism: our experience and systematic review. Pituitary 25, 444–453 (2022). https://doi.org/10.1007/s11102-022-01209-z
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11102-022-01209-z