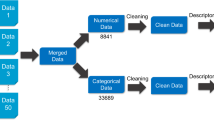
Abstract
Objective
This study aimed to improve the efficiency of pharmacotherapy for CNS diseases by optimizing the ability of drug molecules to penetrate the Blood-Brain Barrier (BBB).
Methods
We established qualitative and quantitative databases of the ADME properties of drugs and derived characteristic features of compounds with efficient BBB penetration. Using these insights, we developed four machine learning models to predict a drug's BBB permeability by assessing ADME properties and molecular topology. We then validated the models using the B3DB database. For acyclovir and ceftriaxone, we modified the Hydrogen Bond Donors and Acceptors, and evaluated the BBB permeability using the predictive model.
Results
The machine learning models performed well in predicting BBB permeability on both internal and external validation sets. Reducing the number of Hydrogen Bond Donors and Acceptors generally improves BBB permeability. Modification only enhanced BBB penetration in the case of acyclovir and not ceftriaxone.
Conclusions
The machine learning models developed can accurately predict BBB permeability, and many drug molecules are likely to have increased BBB penetration if the number of Hydrogen Bond Donors and Acceptors are reduced. These findings suggest that molecular modifications can enhance the efficacy of CNS drugs and provide practical strategies for drug design and development. This is particularly relevant for improving drug penetration of the BBB.
Graphical Abstract







Similar content being viewed by others
Data Availability
This study utilized a database of 307 compounds with quantitative BBB permeability descriptions, which is available in Supplementary Material (1). Additionally, a database of 494 CNS drugs was used, which can be found in Supplementary Material (2). Furthermore, the substructures obtained by fragmenting acyclovir using the Morgan algorithm, as well as the predicted results for the B3DB database, are also provided in Supplementary Material (3) and (4), respectively. The training script used in this study can be obtained by contacting the corresponding author.
References
Daneman R, Prat A. The blood-brain barrier. Cold Spring Harb Perspect Biol. 2015;7(1):a020412. https://doi.org/10.1101/cshperspect.a020412.
Achar A, Myers R, Ghosh C. Drug Delivery Challenges in Brain Disorders across the Blood-Brain Barrier: Novel Methods and Future Considerations for Improved Therapy. Biomedicines. 2021;9(12):1834. https://doi.org/10.3390/biomedicines9121834.
Dong X. Current Strategies for Brain Drug Delivery. Theranostics. 2018;8(6):1481–93. https://doi.org/10.7150/thno.21254.
Wu D, Chen Q, Chen X, et al. The blood-brain barrier: structure, regulation, and drug delivery. Signal Transd Target Ther. 2023;(1):217.
Saxena D, Sharma A, Siddiqui MH, Kumar R. Blood Brain Barrier Permeability Prediction Using Machine Learning Techniques: An Update. Curr Pharm Biotechnol. 2019;20(14):1163–71. https://doi.org/10.2174/1389201020666190821145346.
Banks WA. From blood-brain barrier to blood-brain interface: new opportunities for CNS drug delivery. Nat Rev Drug Discov. 2016;15(4):275–92. https://doi.org/10.1038/nrd.2015.21.
Diéguez-Santana K, Casañola-Martin GM, Torres R, Rasulev B, Green JR, González-Díaz H. Machine Learning Study of Metabolic Networks vs ChEMBL Data of Antibacterial Compounds. Mol Pharm. 2022;19(7):2151–63. https://doi.org/10.1021/acs.molpharmaceut.2c00029.
Cornelissen FMG, Markert G, Deutsch G, et al. Explaining Blood-Brain Barrier Permeability of Small Molecules by Integrated Analysis of Different Transport Mechanisms. J Med Chem. 2023;66(11):7253–67. https://doi.org/10.1021/acs.jmedchem.2c01824.
Freeman R, Noronha A, Woods J. Next generation phenotyping with quantitative narration for DEGCAGS syndrome. Am J Med Genet A. 2023;(4):1020-1025.
Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. https://doi.org/10.1186/1758-2946-6-13.
Ahmad I, Kuznetsov AE, Pirzada AS, Alsharif KF, Daglia M, Khan H. Computational pharmacology and computational chemistry of 4-hydroxyisoleucine: Physicochemical, pharmacokinetic, and DFT-based approaches. Front Chem. 2023;11:1145974. https://doi.org/10.3389/fchem.2023.1145974.
Kenny PW. Hydrogen-Bond Donors in Drug Design. J Med Chem. 2022;65(21):14261–75. https://doi.org/10.1021/acs.jmedchem.2c01147.
Liang L, Liu Y, Kang B, et al. Large-scale comparison of machine learning algorithms for target prediction of natural products. Brief Bioinform. 2022;23(5):bbac359. https://doi.org/10.1093/bib/bbac359.
Kashou AH, May AM, Noseworthy PA. Comparison of two artificial intelligence-augmented ECG approaches: Machine learning and deep learning. J Electrocardiol. 2023;79:75–80. https://doi.org/10.1016/j.jelectrocard.2023.03.009.
Hosseinzadeh A, Zhou JL, Altaee A, et al. Machine learning modeling and analysis of biohydrogen production from wastewater by dark fermentation process. Bioresour Technol. 2021;126111.
Khurshid H, Mumtaz R, Alvi N, et al. Bacterial prediction using internet of things (IoT) and machine learning. Environ Monit Assess. 2022;194(2):133. https://doi.org/10.1007/s10661-021-09698-4.
Meng F, Xi Y, Huang J, Ayers PW. A curated diverse molecular database of blood-brain barrier permeability with chemical descriptors. Sci Data. 2021;8(1):289. https://doi.org/10.1038/s41597-021-01069-5.
Wei CN, Wang LY, Chang XY, Zhou QH. A prediction model using machine-learning algorithm for assessing intrathecal hyperbaric bupivacaine dose during cesarean section. BMC Anesthesiol. 2021;21(1):116. https://doi.org/10.1186/s12871-021-01331-8.
Zhang M, Zhang Y, Yu S, et al. Two machine learning approaches for predicting cyanobacteria abundance in aquaculture ponds. Ecotoxicol Environ Saf. 2023;258:114944. https://doi.org/10.1016/j.ecoenv.2023.114944.
Mondol SIMMR, Kim HJ, Kim KS, Lee S. Machine Learning-Based Hearing Aid Fitting Personalization Using Clinical Fitting Data. J Healthc Eng. 2022;2022:1667672. https://doi.org/10.1155/2022/1667672.
Geldenhuys WJ, Mohammad AS, Adkins CE, Lockman PR. Molecular determinants of blood-brain barrier permeation. Ther Deliv. 2015;6(8):961–71. https://doi.org/10.4155/tde.15.32.
Ballatore C, Huryn DM, Smith AB 3rd. Carboxylic acid (bio)isosteres in drug design. ChemMedChem. 2013;8(3):385–95. https://doi.org/10.1002/cmdc.201200585.
Sun S, Jia Q, Zhang Z. Applications of amide isosteres in medicinal chemistry. Bioorg Med Chem Lett. 2019;29(18):2535–50. https://doi.org/10.1016/j.bmcl.2019.07.033.
Kadela-Tomanek M, Jastrzębska M, Marciniec K, Chrobak E, Bębenek E, Boryczka S. Lipophilicity, Pharmacokinetic Properties, and Molecular Docking Study on SARS-CoV-2 Target for Betulin Triazole Derivatives with Attached 1,4-Quinone. Pharmaceutics. 2021;13(6):781. https://doi.org/10.3390/pharmaceutics13060781.
Li X, Romero MD, Tcaturian S, Kurpiewska K, Dömling A. N-Edited Guanine Isosteres. J Org Chem. 2023;88(14):9823–34. https://doi.org/10.1021/acs.joc.3c00467.
Sun D, Gao W, Hu H, Zhou S. Why 90% of clinical drug development fails and how to improve it? Acta Pharm Sin B. 2022;12(7):3049–62. https://doi.org/10.1016/j.apsb.2022.02.002.
Fan J, Yang J, Jiang Z. Prediction of Central Nervous System Side Effects Through Drug Permeability to Blood-Brain Barrier and Recommendation Algorithm. J Comput Biol. 2018;25(4):435–43. https://doi.org/10.1089/cmb.2017.0149.
Almutairi MM, Gong C, Xu YG, Chang Y, Shi H. Factors controlling permeability of the blood-brain barrier. Cell Mol Life Sci. 2016;73(1):57–77. https://doi.org/10.1007/s00018-015-2050-8.
Dankbaar JW, Hom J, Schneider T, et al. Age- and anatomy-related values of blood-brain barrier permeability measured by perfusion-CT in non-stroke patients. J Neuroradiol. 2009;36(4):219–27. https://doi.org/10.1016/j.neurad.2009.01.001.
Bernardo-Castro S, Sousa JA, Brás A, et al. Pathophysiology of Blood-Brain Barrier Permeability Throughout the Different Stages of Ischemic Stroke and Its Implication on Hemorrhagic Transformation and Recovery. Front Neurol. 2020;11:594672. https://doi.org/10.3389/fneur.2020.594672.
Acknowledgements
This work was supposed by Operation Subsidy Project of Guangxi Key Laboratory of TCM Efficacy Research in 2020 (Grant No: 20-065-38).
Funding
Guangxi Key Laboratory of TCM Efficacy Research in 2020,20-065-38, Jiagang Deng
Author information
Authors and Affiliations
Contributions
Qi Yang proposed innovative ideas and wrote the first draft and code of this article. Lili Fan was responsible for review and revision, Erwei Hao, Xiaotao Hou, Zhengcai Du, and Zhongshang Xia were responsible for collecting data and providing resources, and Jiagang Deng was responsible for providing project support. All authors finally reviewed this article and agreed to submit it.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yang, Q., Fan, L., Hao, E. et al. Machine Learning Exploration of the Relationship Between Drugs and the Blood–Brain Barrier: Guiding Molecular Modification. Pharm Res (2024). https://doi.org/10.1007/s11095-024-03686-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11095-024-03686-2