Abstract
Background
Gastric cancer (GC) remains a significant health problem and carries with it substantial morbidity and mortality. Chidamide is a novel and orally administered histone deacetylase (HDAC) inhibitor and has been demonstrated its anti-tumor efficacy on different kinds of hematological and solid tumors. However, the underlying mechanism of chidamide resistance is still poorly characterized.
Methods
We established chidamide resistant GC cell lines, AGS ChiR and MGC803 ChiR and investigated the toxicologic effects through cell survival, colony formation and flow cytometry assays in vitro, and a subcutaneous xenograft model in vivo. RNA-sequence was then performed to screen chidamide resistance-associated genes between AGS and AGS ChiR cells. The role of Lymphocyte cytosolic protein 1 (LCP1) in chidamide resistance was explored by gain- and loss-of-function analyses.
Results
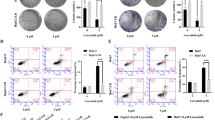
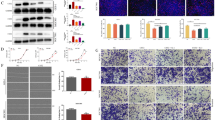
We found that chidamide significantly inhibited cell proliferation and induced the apoptosis in a concentration-dependent manner in wild-type GC cell lines as compared to chidamide resistant cell lines. The transcriptomic profiling, quantitative RT-PCR, and western blot data revealed that LCP1 was upregulated in AGS ChiR cells compared with parental cells. Overexpression of LCP1 conferred and knockdown of LCP1 attenuated the chidamide resistance of GC cells. Epigenetic derepression of LCP1 by chidamide may be a possible reason for the contribution of LCP1 to chidamide resistance.
Conclusions
These findings illustrated that LCP1 may play a chidamide resistance role in GC, suggesting that LCP1 could be a potential target for the therapy of GC combined with chidamide.





Similar content being viewed by others
Abbreviations
- CFDA:
-
China Food and Drug Administration
- DMEM:
-
Dulbecco’s Modified Eagle Medium
- DMSO:
-
Dimethyl sulfoxide
- DEG:
-
Differentially expressed gene
- FBS:
-
Fetal bovine serum
- FDA:
-
Food and Drug Administration
- GC:
-
Gastric cancer
- HCC:
-
Hepatocellular carcinoma
- HDACi:
-
Histone deacetylase inhibitor
- LCP1:
-
Lymphocyte cytosolic protein 1
- NSCLC:
-
Non-Small-Cell Lung carcinoma
- PC:
-
Pancreatic cancer (PC)
- TSA:
-
Trichostatin A
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, et al. Cancer statistics in China, 2015. CA: a cancer journal for clinicians. 2016;66(2):115–32. https://doi.org/10.3322/caac.21338.
Eusebi LH, Telese A, Marasco G, Bazzoli F, Zagari RM. Gastric cancer prevention strategies: A global perspective. J Gastroenterol Hepatol. 2020;35(9):1495–502. https://doi.org/10.1111/jgh.15037.
Smyth EC, Nilsson M, Grabsch HI, van Grieken NC, Lordick F. Gastric cancer. Lancet. 2020;396(10251):635–48. https://doi.org/10.1016/S0140-6736(20)31288-5.
Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92(15):1210–6.
Shah RR. Safety and Tolerability of Histone Deacetylase (HDAC) Inhibitors in Oncology. Drug Saf. 2019;42(2):235–45. https://doi.org/10.1007/s40264-018-0773-9.
San-Miguel JF, Hungria VTM, Yoon S-S, Beksac M, Dimopoulos MA, Elghandour A, et al. Panobinostat plus bortezomib and dexamethasone versus placebo plus bortezomib and dexamethasone in patients with relapsed or relapsed and refractory multiple myeloma: a multicentre, randomised, double-blind phase 3 trial. Lancet Oncol. 2014;15(11):1195–206. https://doi.org/10.1016/S1470-2045(14)70440-1.
Ning Z-Q, Li Z-B, Newman MJ, Shan S, Wang X-H, Pan D-S, et al. Chidamide (CS055/HBI-8000): a new histone deacetylase inhibitor of the benzamide class with antitumor activity and the ability to enhance immune cell-mediated tumor cell cytotoxicity. Cancer Chemother Pharmacol. 2012;69(4):901–9. https://doi.org/10.1007/s00280-011-1766-x.
Dong M, Ning ZQ, Xing PY, Xu JL, Cao HX, Dou GF, et al. Phase I study of chidamide (CS055/HBI-8000), a new histone deacetylase inhibitor, in patients with advanced solid tumors and lymphomas. Cancer Chemother Pharmacol. 2012;69(6):1413–22. https://doi.org/10.1007/s00280-012-1847-5.
Liu L, Qiu S, Liu Y, Liu Z, Zheng Y, Su X, et al. Chidamide and 5-flurouracil show a synergistic antitumor effect on human colon cancer xenografts in nude mice. Neoplasma. 2016;63(2):193–200. https://doi.org/10.4149/203_150422N214.
Chan TS, Tse E, Kwong Y-L. Chidamide in the treatment of peripheral T-cell lymphoma. Onco Targets Ther. 2017;10:347–52. https://doi.org/10.2147/OTT.S93528.
Lu X, Ning Z, Li Z, Cao H, Wang X. Development of chidamide for peripheral T-cell lymphoma, the first orphan drug approved in China. Intractable Rare Dis Res. 2016;5(3):185–91. https://doi.org/10.5582/irdr.2016.01024.
Zhou Y, Pan D-S, Shan S, Zhu J-Z, Zhang K, Yue X-P, et al. Non-toxic dose chidamide synergistically enhances platinum-induced DNA damage responses and apoptosis in Non-Small-Cell lung cancer cells. Biomed Pharmacother. 2014;68(4):483–91. https://doi.org/10.1016/j.biopha.2014.03.011.
Wang H, Guo Y, Fu M, Liang X, Zhang X, Wang R, et al. Antitumor activity of Chidamide in hepatocellular carcinoma cell lines. Mol Med Rep. 2012;5(6):1503–8. https://doi.org/10.3892/mmr.2012.858.
Qiao Z, Ren S, Li W, Wang X, He M, Guo Y, et al. Chidamide, a novel histone deacetylase inhibitor, synergistically enhances gemcitabine cytotoxicity in pancreatic cancer cells. Biochem Biophys Res Commun. 2013;434(1). https://doi.org/10.1016/j.bbrc.2013.03.059.
Zhang W, Niu J, Ma Y, Yang X, Cao H, Guo H, et al. The Synergistic Antitumor Activity of Chidamide in Combination with Bortezomib on Gastric Cancer. Onco Targets Ther. 2020;13:3823–37. https://doi.org/10.2147/OTT.S240721.
Luo Se, Ma K, Zhu H, Wang S, Liu M, Zhang W, et al. Molecular, biological characterization and drug sensitivity of chidamide-resistant non-small cell lung cancer cells. Oncol Lett. 2017;14(6):6869–75. https://doi.org/10.3892/ol.2017.7060.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20. https://doi.org/10.1093/bioinformatics/btu170.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–60. https://doi.org/10.1038/nmeth.3317.
Roberts A, Trapnell C, Donaghey J, Rinn JL, Pachter L. Improving RNA-Seq expression estimates by correcting for fragment bias. Genome Biol. 2011;12(3):R22. https://doi.org/10.1186/gb-2011-12-3-r22.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28(5):511–5. https://doi.org/10.1038/nbt.1621.
Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. https://doi.org/10.1093/bioinformatics/btu638.
Anders S, Huber W. Differential expression of RNA-Seq data at the gene level – the DESeq package. EMBL. 2013.
Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36(Database issue):D480-D4.
Zeng Q, Li L, Feng Z, Luo L, Xiong J, Jie Z, et al. LCP1 is a prognostic biomarker correlated with immune infiltrates in gastric cancer. Cancer Biomark. 2021;30(1):105–25. https://doi.org/10.3233/CBM-200006.
Zhu Y, Das K, Wu J, Lee MH, Tan P. RNH1 regulation of reactive oxygen species contributes to histone deacetylase inhibitor resistance in gastric cancer cells. Oncogene. 2014;33(12):1527–37. https://doi.org/10.1038/onc.2013.104.
Dedes KJ, Dedes I, Imesch P, von Bueren AO, Fink D, Fedier A. Acquired vorinostat resistance shows partial cross-resistance to “second-generation” HDAC inhibitors and correlates with loss of histone acetylation and apoptosis but not with altered HDAC and HAT activities. Anticancer Drugs. 2009;20(5):321–33. https://doi.org/10.1097/CAD.0b013e3283262a32.
Yamada H, Arakawa Y, Saito S, Agawa M, Kano Y, Horiguchi-Yamada J. Depsipeptide-resistant KU812 cells show reversible P-glycoprotein expression, hyper-acetylated histones, and modulated gene expression profile. Leuk Res. 2006;30(6):723–34.
Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Genet. 2019;20(11):631–56. https://doi.org/10.1038/s41576-019-0150-2.
Goldstein D, Djeu J, Latter G, Burbeck S, Leavitt J. Abundant synthesis of the transformation-induced protein of neoplastic human fibroblasts, plastin, in normal lymphocytes. Cancer Res. 1985;45(11 Pt 2):5643–7.
Koide N, Kasamatsu A, Endo-Sakamoto Y, Ishida S, Shimizu T, Kimura Y, et al. Evidence for Critical Role of Lymphocyte Cytosolic Protein 1 in Oral Cancer. Sci Rep. 2017;7:43379. https://doi.org/10.1038/srep43379.
Foran E, McWilliam P, Kelleher D, Croke DT, Long A. The leukocyte protein L-plastin induces proliferation, invasion and loss of E-cadherin expression in colon cancer cells. Int J Cancer. 2006;118(8):2098–104.
Su Kim D, Choi YD, Moon M, Kang S, Lim J-B, Kim KM, et al. Composite three-marker assay for early detection of kidney cancer. Cancer Epidemiol Biomarkers Prev. 2013;22(3):390–8. https://doi.org/10.1158/1055-9965.EPI-12-1156.
Luo W, Schork NJ, Marschke KB, Ng S-C, Hermann TW, Zhang J, et al. Identification of polymorphisms associated with hypertriglyceridemia and prolonged survival induced by bexarotene in treating non-small cell lung cancer. Anticancer Res. 2011;31(6):2303–11.
Janji B, Vallar L, Al Tanoury Z, Bernardin F, Vetter G, Schaffner-Reckinger E, et al. The actin filament cross-linker L-plastin confers resistance to TNF-alpha in MCF-7 breast cancer cells in a phosphorylation-dependent manner. J Cell Mol Med. 2010;14(6A):1264–75. https://doi.org/10.1111/j.1582-4934.2009.00918.x.
ACKNOWLEGMENTS AND DISCLOSURES
The authors have declared that they have no competing interests.
Funding
This study was supported by grants from the National Natural Science Foundation of China (No.82160468), Shanghai Science and Technology Innovation Action Plan, Medical Innovation Research Special Project (No.20Y11908400) and The Top-level Clinical Discipline Project of Shanghai Pudong (No. PWYgf2021-07).
Author information
Authors and Affiliations
Contributions
Conception and design: Jingde Chen and Yong Gao. Conduction of experiments: Wenfang Bao, Zhe Zhu. Statical analysis and interpretation: Wenfang Bao, Zhe Zhu. Original manuscript drafting and figure construction: Wenfang Bao and Zhe Zhu. Manuscript editing and completion: Jingde Chen. All authors contributed to the article and approved the submitted version.
Corresponding authors
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bao, W., Zhu, Z., Gao, Y. et al. Transcriptome Profiling Analysis Identifies LCP1 as a Contributor for Chidamide Resistance in Gastric Cancer. Pharm Res 39, 867–876 (2022). https://doi.org/10.1007/s11095-022-03291-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11095-022-03291-1