Abstract
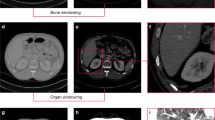
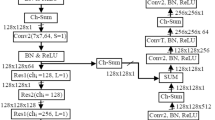
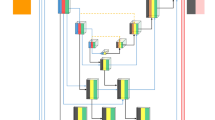
Nanotechnology and photonics, two of the most promising fields of the 21st century, come together in nanophotonics. Its primary advantage is that it may be used to build a number of innovative features based on local electromagnetic interaction. Computed tomography (CT) imaging considerably aids in the identification, prognosis, and assessment of therapy response in kidney cancer. This paper presents a unique approach for nuclear image-based kidney tumour identification using deep learning-based segmentation and classification. In addition to CT-features-based gene mutation identification, segmentation-free volume estimate, autonomous kidney localization, and diagnosis of cancer, we developed deep learning approaches. The plan is to boost the contrast of the picture using a convolutional network-based active contour normalisation and then categorise the segmented image with a stacked Monte Carlo Markov encoder neural network. In the experimental study, we look at how different sets of nuclear images perform in terms of training accuracy, Jaccard index, root mean square error (RMSE), NSE, average precision, and F-measure. Training accuracy of 95% was achieved using the suggested method, along with a Jaccard index of 62%, RMSE of 58%, NSE of 61%, average precision of 65%, and F-measure of 71%.











Similar content being viewed by others
Data and materials availability
All the data’s available in the manuscript.
References
Abdelrahman, A., Viriri, S.: Kidney tumor semantic segmentation using deep learning: a survey of state-of-the-art. J. Imaging 8(3), 55 (2022)
Abdeltawab, H.A., Khalifa, F.A., Ghazal, M.A., Cheng, L., El-Baz, A.S., Gondim, D.D.: A deep learning framework for automated classification of histopathological kidney whole-slide images. J. Pathol. Inf. 13, 100093 (2022)
Abu Haeyeh, Y., Ghazal, M., El-Baz, A., Talaat, I.M.: Development and evaluation of a novel deep-learning-based framework for the classification of renal histopathology images. Bioengineering 9(9), 423 (2022)
Alzu’bi, D., Abdullah, M., Hmeidi, I., AlAzab, R., Gharaibeh, M., El-Heis, M., Abualigah, L.: Kidney tumor detection and classification based on deep learning approaches: a new dataset in CT scans. J. Healthc. Eng. (2022). https://doi.org/10.1155/2022/3861161
Gharaibeh, M., Alzu’bi, D., Abdullah, M., Hmeidi, I., Al Nasar, M.R., Abualigah, L., Gandomi, A.H.: Radiology imaging scans for early diagnosis of kidney tumors: a review of data analytics-based machine learning and deep learning approaches. Big Data Cogn. Comput. 6(1), 29 (2022)
Hao, F., Liu, X., Li, M., Han, W.: Accurate kidney pathological image classification method based on deep learning and multi-modal fusion method with application to membranous nephropathy. Life 13(2), 399 (2023)
Hsiao, C.H., Sun, T.L., Lin, P.C., Peng, T.Y., Chen, Y.H., Cheng, C.Y., Huang, Y.: A deep learning-based precision volume calculation approach for kidney and tumor segmentation on computed tomography images. Comput. Method Programs Biomed. 221, 106861 (2022a)
Hsiao, C.H., Lin, P.C., Chung, L.A., Lin, F.Y.S., Yang, F.J., Yang, S.Y., Sun, T.L.: A deep learning-based precision and automatic kidney segmentation system using efficient feature pyramid networks in computed tomography images. Comput. Method Programs Biomed. 221, 106854 (2022b)
Kang, L., Zhou, Z., Huang, J., Han, W., Member, I.E.E.E.: Renal tumors segmentation in abdomen CT Images using 3D-CNN and ConvLSTM. Biomed. Signal Process. Control 72, 103334 (2022)
Kers, J., Bülow, R.D., Klinkhammer, B.M., Breimer, G.E., Fontana, F., Abiola, A.A., Boor, P.: Deep learning-based classification of kidney transplant pathology: a retrospective, multicentre, proof-of-concept study. Lancet Digit. Health 4(1), e18–e26 (2022)
Kong, J., He, Y., Zhu, X., Shao, P., Xu, Y., Chen, Y., Yang, G.: BKC-Net: Bi-knowledge contrastive learning for renal tumor diagnosis on 3D CT images. Knowl.-Based Syst. 252, 109369 (2022)
Liu, J., Yildirim, O., Akin, O., Tian, Y.: AI-driven robust kidney and renal mass segmentation and classification on 3D CT images. Bioengineering 10(1), 116 (2023)
Murugesan, M., Kaliannan, K., Balraj, S., Singaram, K., Kaliannan, T., Albert, J.R.: A hybrid deep learning model for effective segmentation and classification of lung nodules from CT images. J. Intell. Fuzzy Syst. 42(3), 2667–2679 (2022)
Nasir, M.U., Zubair, M., Ghazal, T.M., Khan, M.F., Ahmad, M., Rahman, A.U., Mansoor, W.: Kidney cancer prediction empowered with blockchain security using transfer learning. Sensors 22(19), 7483 (2022)
Robinson-Weiss, C., Patel, J., Bizzo, B.C., Glazer, D.I., Bridge, C.P., Andriole, K.P., Mayo-Smith, W.W.: Machine learning for adrenal gland segmentation and classification of normal and adrenal masses at CT. Radiology 306, e220101 (2022)
Roblot, V., Giret, Y., Mezghani, S., Auclin, E., Arnoux, A., Oudard, S., Fournier, L.: Validation of a deep learning segmentation algorithm to quantify the skeletal muscle index and sarcopenia in metastatic renal carcinoma. Eur. Radiol. 32(7), 4728–4737 (2022)
Saxena, S.K., Shrivastava, J.N., Agarwal, G., Kumar, S.: Classification of kidney diseases using transfer learning. In: Handbook of research on applications of AI, digital twin, and internet of things for sustainable development, pp. 47–60. IGI Global (2023).
Srinivasan, M., Soji, E.S.: Kidney tumour segmentation and classification using deep learning. Cent. Asian J. Med. Nat. Sci. 3(6), 247–269 (2022)
Xu, Q., Zhu, Q., Liu, H., Chang, L., Duan, S., Dou, W., Ye, J.: Differentiating benign from malignant renal tumors using T2-and diffusion-weighted images: a comparison of deep learning and radiomics models versus assessment from radiologists. J. Magn. Resonance Imaging 55(4), 1251–1259 (2022)
Zhou, T., Guan, J., Feng, B., Xue, H., Cui, J., Kuang, Q., Long, W.: Distinguishing common renal cell carcinomas from benign renal tumors based on machine learning: comparing various CT imaging phases, slices, tumor sizes, and ROI segmentation strategies. Eur. Radiol. (2023). https://doi.org/10.1007/s00330-022-09384-0
Zhu, X.L., Shen, H.B., Sun, H., Duan, L.X., Xu, Y.Y.: Improving segmentation and classification of renal tumors in small sample 3D CT images using transfer learning with convolutional neural networks. Int. J. Comput. Assist. Radiol. Surg. 17(7), 1303–1311 (2022)
Funding
This research not received any fund.
Author information
Authors and Affiliations
Contributions
M.M. Conceived and design the analysis Writing- Original draft preparation. N.K. Collecting the Data, V.V. Contributed data and analysis stools S.G. Performed and analysis, A.K.P. Performed and analysis, S.G. Wrote the Paper R.B. Editing and Figure Design.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
This article does not contain any studies with animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Manjula, M., Kumar, N., Vekariya, V. et al. Nanoparticle analysis based on optical ion beam in nuclear imaging by deep learning architectures. Opt Quant Electron 55, 863 (2023). https://doi.org/10.1007/s11082-023-05141-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11082-023-05141-9