Abstract
Purpose
Computed tomography (CT) images can display internal organs of patients and are particularly suitable for preoperative surgical diagnoses. The increasing demands for computer-aided systems in recent years have facilitated the development of many automated algorithms, especially deep convolutional neural networks, to segment organs and tumors or identify diseases from CT images. However, performances of some systems are highly affected by the amount of training data, while the sizes of medical image data sets, especially three-dimensional (3D) data sets, are usually small. This condition limits the application of deep learning.
Methods
In this study, given a practical clinical data set that has 3D CT images of 20 patients with renal carcinoma, we designed a pipeline employing transfer learning to alleviate the detrimental effect of the small sample size. A dual-channel fine segmentation network (FS-Net) was constructed to segment kidney and tumor regions, with 210 publicly available 3D images from a competition employed during the training phase. We also built discriminative classifiers to classify the benign and malignant tumors based on the segmented regions, where both handcrafted and deep features were tested.
Results
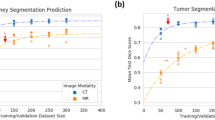
Our experimental results showed that the Dice values of segmented kidney and tumor regions were 0.9662 and 0.7685, respectively, which were better than those of state-of-the-art methods. The classification model using radiomics features can classify most of the tumors correctly.
Conclusions
The designed FS-Net was demonstrated to be more effective than simply fine-tuning on the practical small size data set given that the model can borrow knowledge from large auxiliary data without diluting the signal in primary data. For the small data set, radiomics features outperformed deep features in the classification of benign and malignant tumors. This work highlights the importance of architecture design in transfer learning, and the proposed pipeline is anticipated to provide a reference and inspiration for small data analysis.






Similar content being viewed by others
References
Nie P, Yang G, Wang Z, Yan L, Miao W, Hao D, Wu J, Zhao Y, Gong A, Cui J, Jia Y, Niu H (2020) A CT-based radiomics nomogram for differentiation of renal angiomyolipoma without visible fat from homogeneous clear cell renal cell carcinoma. Eur Radiol 30:1274–1284. https://doi.org/10.1007/s00330-019-06427-x
Badura P, Wieclawek W, Pycinski B (2016) Automatic 3D segmentation of renal cysts in CT. Adv Intell Syst Comput 471:149–163. https://doi.org/10.1007/978-3-319-39796-2_13
Kasinathan G, Jayakumar S, Gandomi AH, Ramachandran M, Fong SJ, Patan R (2019) Automated 3-D lung tumor detection and classification by an active contour model and CNN classifier. Expert Syst Appl 134:112–119. https://doi.org/10.1016/j.eswa.2019.05.041
Lin F, Cui EM, Lei Y, Luo LP (2019) CT-based machine learning model to predict the Fuhrman nuclear grade of clear cell renal cell carcinoma. Abdom Radiol 44:2528–2534. https://doi.org/10.1007/s00261-019-01992-7
Furumoto H, Shimada Y, Imai K, Maehara S, Maeda J, Hagiwara M, Okano T, Masuno R, Kakihana M, Kajiwara N, Ohira T, Ikeda N (2018) Prognostic impact of the integration of volumetric quantification of the solid part of the tumor on 3DCT and FDG-PET imaging in clinical stage IA adenocarcinoma of the lung. Lung Cancer 121:91–96. https://doi.org/10.1016/j.lungcan.2018.05.001
Lin Z, Cui Y, Liu J, Sun Z, Ma S, Zhang X, Wang X (2021) Automated segmentation of kidney and renal mass and automated detection of renal mass in CT urography using 3D U-Net-based deep convolutional neural network. Eur Radiol 31:5021–5031. https://doi.org/10.1007/s00330-020-07608-9
Türk F, Lüy M, Barışçı N (2020) Kidney and renal tumor segmentation using a hybrid v-net-based model. Mathematics 8(10):1772. https://doi.org/10.3390/math8101772
Yang G, Li G, Pan T, Kong Y, Wu J, Shu H, Luo L, Dillenseger JL, Coatrieux JL, Tang L, Zhu X (2018) Automatic segmentation of kidney and renal tumor in CT images based on 3D fully convolutional neural network with pyramid pooling module. In: Proceedings—international conference on pattern recognition. IEEE, pp 3790–3795. https://doi.org/10.1109/ICPR.2018.8545143.
De Perrot T, Hofmeister J, Burgermeister S, Martin SP, Feutry G, Klein J, Montet X (2019) Differentiating kidney stones from phleboliths in unenhanced low-dose computed tomography using radiomics and machine learning. Eur Radiol 29:4776–4782. https://doi.org/10.1007/s00330-019-6004-7
Heller N, Sathianathen N, Kalapara A, Walczak E, Moore K, Kaluzniak H, Rosenberg J, Blake P, Rengel Z, Oestreich M, Dean J, Tradewell M, Shah A, Tejpaul R, Edgerton Z, Peterson M, Raza S, Regmi S, Papanikolopoulos N, Weight C (2019) The KiTS19 Challenge Data: 300 Kidney Tumor Cases with Clinical Context, CT Semantic Segmentations, and Surgical Outcomes. arXiv preprint arXiv:1904.00445
Heller N, Isensee F, Maier-Hein KH, Hou X, Xie C, Li F, Nan Y, Mu G, Lin Z, Han M, Yao G, Gao Y, Zhang Y, Wang Y, Hou F, Yang J, Xiong G, Tian J, Zhong C, Ma J, Rickman J, Dean J, Stai B, Tejpaul R, Oestreich M, Blake P, Kaluzniak H, Raza S, Rosenberg J, Moore K, Walczak E, Rengel Z, Edgerton Z, Vasdev R, Peterson M, McSweeney S, Peterson S, Kalapara A, Sathianathen N, Papanikolopoulos N, Weight C (2021) The state of the art in kidney and kidney tumor segmentation in contrast-enhanced CT imaging: results of the KiTS19 challenge. Med Image Anal 67:101821. https://doi.org/10.1016/j.media.2020.101821
Chen S, Ma K, Zheng Y (2019) Med3D: transfer learning for 3D medical image analysis. arXiv preprint arXiv:1904.00625
Tibshirani R (1996) Regression shrinkage and selection via the Lasso. J R Stat Soc Ser B 58(1):267–288. https://doi.org/10.1111/j.2517-6161.1996.tb02080.x
Wall ME, Rechtsteiner A, Rocha LM (2003) Singular value decomposition and principal component analysis. In: A practical approach to microarray data analysis. Springer, pp 91–109. https://doi.org/10.1007/0-306-47815-3_5
Menze B, Kelm B, Masuch R, Himmelreich U, Bachert P, Petrich W, Hamprecht F (2009) A comparison of Random Forest and its Gini importance with standard chemometric methods for the feature selection and classification of spectral data. BMC Bioinformatics 10:213. https://doi.org/10.1186/1471-2105-10-213
Kononenko I (1994) Estimating attributes: Analysis and extensions of RELIEF. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 784 LNCS:171–182. https://doi.org/10.1007/3-540-57868-4_57
Grabczewski K, Jankowski N (2005) Feature selection with decision tree criterion. In: Fifth International Conference on Hybrid Intelligent Systems (HIS’05). p 6 pp. https://doi.org/10.1109/ICHIS.2005.43
Escanilla NS, Hellerstein L, Kleiman R, Kuang Z, Shull J, Page D (2019) Recursive feature elimination by sensitivity testing. In: Proceedings—17th IEEE international conference on machine learning and applications, ICMLA 2018. pp 40–47. https://doi.org/10.1109/ICMLA.2018.00014
Jin X, Xu A, Bie R, Guo P (2006) Machine learning techniques and chi-square feature selection for cancer classification using SAGE gene expression profiles. In: Lecture Notes in Computer Science (including subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics). Springer, pp 106–115. https://doi.org/10.1007/11691730_11
Isensee F, Jaeger P, Kohl S, Petersen J, Maier-Hein K (2021) nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat Methods 18(2):203–211. https://doi.org/10.1038/s41592-020-01008-z
Mzurikwao D, Khan MU, Samuel OW, Cinatl J Jr, Wass M, Michaelis M, Marcelli G, Ang CS (2020) Towards image-based cancer cell lines authentication using deep neural networks. Sci Rep 10:19857. https://doi.org/10.1038/s41598-020-76670-6
van der Maaten L, Hinton G (2008) Viualizing data using t-SNE. J Mach Learn Res 9:2579–2605
Lee H, Hong H, Park S, Kim J (2017) Differentiation of fat-poor angiomyolipoma from clear cell renal cell carcinoma in contrast-enhanced MDCT images using quantitative feature classification. Med Phys 44(7):3604–3614. https://doi.org/10.1002/mp.12258
Funding
This work was supported in part by the National Natural Science Foundation of China (61803196), Natural Science Foundation of Guangdong Province of China (2020A1515010038), and Presidential Foundation of Zhujiang Hospital of Southern Medical University (yzjj2018rc03).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared that they have no conflicts of interest in this work.
Ethical approval
This work was conducted retrospectively on data from clinical routine which was completely anonymized. Ethical approval was, therefore, not required. This work relies on the KiTS19 data set. For use of these data sets, no ethics statements are necessary.
Informed consent
Informed consent was obtained from all individual participants included in the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, XL., Shen, HB., Sun, H. et al. Improving segmentation and classification of renal tumors in small sample 3D CT images using transfer learning with convolutional neural networks. Int J CARS 17, 1303–1311 (2022). https://doi.org/10.1007/s11548-022-02587-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11548-022-02587-2