Abstract
The synchronization patterns exhibited by small networks of neurons that regulate biological processes (CPGs) have aroused growing scientific interest. In many of these networks there is a main behavioral pattern within the parameter space. In particular, in the context of insect locomotion, tripod walking stands out as a predominant pattern, both in natural observations (where insects walk on tripod gait) and in mathematical models. This predominance appears to be stable under parameter variations within the network, suggesting a possible correlation with the underlying network topology. Tripod walking can be naturally extended to all CPGs with a bipartite connectivity. Then a natural question arises: Are “generalized tripod gaits” equally dominant among synchronization patterns within those networks? To investigate this, we carried out a comprehensive study covering all bipartite networks of up to nine neurons. For each of those networks we numerically explore the phase space using a quasi-MonteCarlo method to see what are the main synchronization patterns that the network can achieve. Then, all those patterns are grouped according to their dynamics. Generalized tripod gait was observed in all cases examined as the dominant pattern again. However, certain cases revealed additional stable patterns, mainly associated with the 3-colorings of the respective graph structures.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
The study of patterns in directed networks is today a current line of research [1] due to the large number of practical applications. In the large amount of possible network configurations, the bipartite networks are a common type of networks whose nodes are organized into two main groups. Therefore, bipartite networks are useful for pairwise relationships between two different groups of nodes [2], as in predator/pray, producer/consumer, left-side/right-side, and so on. These kinds of situations are quite common in nature, biology, social relationships, food web, ...
The review [3] provides a description of certain important classes of biological networks that exhibit a native bipartite structure and their associated data. Ecological networks are a typical example and can be subdivided into three main types: food webs, mutualistic webs, and host-parasitoid webs. Most of them can be described using bipartite networks [4, 5]. Beyond ecological networks, bipartite biomedical and biomolecular networks, such as gene-disease network [6] and modeling of protein complexes as networks, are more abstract since the network is usually designed using various indicators of human diseases or molecular interactions. Epidemiological networks are another common type of models that use bipartite networks. In this case the nodes are related with individual patients: bipartite structures can be built based on individuals who are classified by gender, location, infectious agent,... Related with these networks are global networks of social relations based primarily on behavioral patterns of individuals or groups.
Recently, the use of more specific models is growing, especially in the study of small neuron networks [7], like the Central Pattern Generators (CPGs), which are neuronal circuits that when activated can generate rhythmic motor patterns even in absence of sensory input (see [8,9,10]).
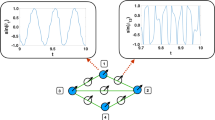
In such networks, structure can be defined at two levels: (a) at the structural connectivity level (how neurons are connected with each others), (b) at the functional connectivity level (how subgroups of neurons are firing in synchrony). At the structural connectivity level, some connectivities are such that they can be represented as exclusive subsets of neurons with connections only among subsets, and no connections among neurons within a subset. When connections are between two subsets only, we will call it a bipartite connectivity (see Fig. 1a where black white circles form the two structural bipartition). At the functional connectivity level, some networks yield rhythmical activities where some subgroups of neurons fire together in turn, so neurons can be now partitioned according to the dynamical pattern of activity (see Fig. 1). When there are two subgroups of synchronized neurons, we will denote this bipartite (dynamical) pattern, and when there are three, we will denote this tripartite (dynamical) pattern.
Hence, in CPGs, the existence of bipartite connectivity is of special interest with regards to their relation with bipartite patterns, like in the CPG that controls the movement of animals [11, 12]. Therefore, bipartite networks can be applied to a wide range of problems, and so a theoretical study of possible small bipartite networks that can lead to some special pattern configurations is of great interest.
A first common attempt to study the dynamics of a network is to consider networks of oscillators [13,14,15], which allows to provide some theoretical insights. In the case of generic small networks we have recently proposed [16] two numerical techniques to deal with small neuron CPGs. In [16, 17] we applied these techniques to the bursting neuron CPG model introduced by Ghigliazza and Holmes [18] to model the movement of insects (cockroaches). Throughout this article, we will call GH network this network.
Tripod gait. In the GH neuron network, with graph structure as in (a), each neuron is assumed to control one of the hexapod’s legs, as it is suggested in (b), where active neurons are painted in red. A leg is moving if its control neuron is active (bursting). During tripod gait, neurons’ bursting intervals synchronize with each others as depicted in (c). Activation and inactivation intervals of neurons are usually represented schematically in an hexagram as the one in (d). The neurons activate in groups of three, i.e., legs move alternatively in groups of three and there are always three or six legs on the ground. The synchronization pattern is associated with the 2-coloring of the graph depicted in (a)
Tetrapod gait. Neurons activate in pairs. Legs move cyclically in three groups of two. This synchronization pattern is associated with the 3-coloring of the graph depicted in (a). Changing one of the parameters of the system (8), the network changes its pattern to the tetrapod gait. See [16] for details
GH network has a bipartite connectivity and can display both bipartite and tripartite patterns, depending on the parameters of the system, showing synchronization patterns that correspond to insect gaits that can be observed in nature, as the tripod (Fig. 1) or the tetrapod gaits (Fig. 2) [16]. In particular, the tripod gait appears as the dominant pattern [19, 20] in a wide region of the parameter space [17]. Since the connections in the model are mutually inhibitory, it is natural to expect that the connected neurons should not all be active at the same time, and so that synchronization pattern would determine a partition of the set of neurons. It is also natural to wonder if the prevalence of tripod gait is to some extent a consequence of network bipartiteness.
The main goal of this article is to investigate if bipartite patterns are dominant in bipartite networks as in the GH network. To see at which extend network connectivity determines the synchronization patterns, we have considered every network made of 6 to 9 neurons with bipartite connectivity. For each of them, we have sampled initial conditions to describe the distribution of its emerging synchronization patterns. We report here some results about those distributions.
This article is organized as follows. In Sect. 2 we introduce the equations and methodologies of the model. Section 2.4 is dedicated to giving the definitions of color synchronization patterns. Section 3 presents the results, shows the dominance of bipartite patterns, and illustrates why in some network topologies it is possible to have non-bipartite patterns. Finally, we present our conclusions at the end of the paper.
2 Neuron model, graph neuron networks and methodologies
2.1 Neuron model
In this article we construct small neuron networks where each neuron follows a Hodgkin-Huxley formalism adapted to the movement of insects (cockroaches [18]).
The dynamics of one isolated neuron are described in terms of a fast nonlinear calcium current, \(I_{Ca}\), a slower potassium current \(I_K\), a very slow current \(I_{KS}\), a linear leakage current \(I_L\), and an external current \(I_\text {ext}\). The ODE system describing the dynamics of the action potential v, the potassium gate m and the slow potassium gate w is
with the auxiliary ionic current functions defined by
and where the different time scales and steady state gating variables are
and
The parameters C, \(\epsilon \) and \(\delta \) determine the time scales of v, m and w. If X denotes any of the considered ions, \(E_X\) is the Nernst potential, \(g_X\) is the maximal conductance, and \(k^0_X\) is the steepness of the transition happening at threshold potential \(v^\text {th}_X\). This model can produce alternation between a rest state with a flat potential and a burst of fast oscillations of v around a high voltage, mimicking the action potential of a neuron without explicit refractory periods nor activation thresholds (as in LIF or QIF neurons). See §3.3 and §4 of [21] for details on its parameters and global dynamics.
The results of these experiments are expected to be similar for other types of excitable neuron models.
2.2 Neuron networks
Consider an undirected, connected and without loops graph with N vertices. We denote the degree of one of its vertices x, the number of adjacent vertices, by \(d_x\). To build its associated neuron network model we consider a copy of (1) for each vertex x which represents a neuron of the network. For any variable “A” of the ODE system (1) the subscripted term “\(A_x\)” denotes the copy of this variable for the neuron x.
Since synapses are directional, we will consider each edge as two opposed synapses between the neurons. We modeled them using the voltage based synapse model of [18], given by the following ODE system
This new synapse variable \(s_x\) of neuron x will affect its post-synaptic neurons as follows: In this paper all synapses are inhibitory, thus, we add a subtractive (inhibitory) term to the first equation of (1) getting
being
where y runs over all the vertices of the graph adjacent to x (cf. [16, 18]) and \(g_\text {syn}\) denotes the synaptic strength. As we can see in (7), we average the action of all pre-synaptic neurons. Hence, it can be seen that the maximal inhibitory current is the same across the network.
For instance, in the case of the GH network we have that the different values of the extra current \((I_\text {syn})_x\) for each neuron potential \(v_x\) are:
In general, with this construction, we obtain a 4N-dimensional ODE system. Obviously, this ODE system presents the same symmetries as the underlying network.
Notice that the inhibitory actions along edges can be asymmetric: the inhibitory current that x receives from y could be different from the inhibitory current that y receives from x, as those are averaged among number of neighbors as in the example of the GH network.
On the other hand, while all the neuron networks are directed (as synapses are), for the sake of simplicity, we just show the topology of the connection graph identifying opposed synapses.
Since we are investigating the ubiquity of a generalization of the tripod gait for other neuron networks, we have taken a set of parameter values for which the tripod gait is ubiquitous in the GH network [17]. We have fixed the parameters of the system to
We should remark that the weights of the synaptic variables \(s_i\) used in [17] are different from those of (8). In our previous work we used the conditions of [18] where pre-synaptic \(s_i\) variables are weighted averaged: the weight of the synapses from 1 to 2, 3 to 2, 4 to 5 and 5 to 6 was half of the weight of the others (neurons numbered as in Fig. 1). Despite of this difference, fixed the rest of the parameters, the dynamic patterns are the same for both dynamical systems, namely, the tripod gait of Fig. 1, see [16, 17].
2.3 Quasi-Monte Carlo sweep. Patterns
The main goal of this article is to investigate if bipartite patterns are dominant in bipartite networks as in the 6-neurons GH CPG network. That is, we want to know the fraction of initial conditions producing a bipartite pattern in the long run compared to the rest of patterns, for all bipartite networks with up to 9 neurons. Since the phase space is 4N-dimensional, where N is the number of neurons, it is computationally unfeasible to sweep a region of this space with a (traditional) fixed step sweep. Even the coarsest of the partitions with 2 single points in each dimension of the swept region would require millions of initial conditions to integrate and the result would not be representative of the swept region (they would be just the corners of the 4N-dimensional cube of initial conditions selected). Hence, we have applied the quasi-Monte Carlo sweep technique [16, 17] described below. For each neuron network, this technique combines: (i) the choice of an appropriate set of initial conditions; (ii) a numerical integration of the neuron network ODE system for each of these initial conditions; and (iii) a final post-processing of the solution until we obtain a pattern, which will be a discrete, purely combinatorial object associated with the underlying network.
Now, for each neuron network we select 200 initial conditions (this is true for the rest of the simulations on this paper) using Halton sequences [22], a well-known low discrepancy sequence generator.
The initial membrane potentials are between \({-20.2}\text {mV}\) and \({4.8}\text {mV}\), and the rest of variables are confined to [0, 1].
The system is integrated, using the DOPRI78 integrator (a classical embedded RK method of Dormand and Prince [23, 24]), for \(t \in [0, 100]\) to allow it to eventually reach a periodic orbit. If this is the case, we compute the bursting (active) intervals of each neuron (the duty cycle). To do so, we compute a moving standard deviation along the voltages and declare a neuron active if this standard deviation surpases a given threshold. Then, we build the firing pattern with this information: the period of the orbit is divided into ranges along which the status of the network (the sets of active and inactive neurons) remains constant. The limit points of each of these ranges coincide with the moment of activation or inactivation of one or more neurons, and vice versa: every moment of activation or inactivation of any neuron is a limit point for these ranges.
Since we want to identify dynamical patterns that are similar, independently of the global time scale, we drop the ranges which are too small with respect to the longest one. More precisely, we fix a parameter \(t_\text {small}\in (0,1)\), and a range is discarded if its length is smaller than \(t_\text {small} \cdot T_\text {MAX}\), where \(T_\text {MAX}\) is the greatest length among all ranges. After this filtering, we obtain a sequence of ranges with the status of each of them, and we remove the length of events to group patterns with the same combinatorial behavior. If the network has p neurons \(x_1,\ldots ,x_{p}\), the status of a specific range can be stored in the binary expansion \(a_1\ldots a_p\) with \(a_i=0\) or 1 if \(x_i\) is inactive or active respectively for \(i=1,\ldots ,p\), or equivalently in the number \(s=\sum _{i=1}^{p}a_i 2^{i-1}\) lying between 0 and \(2^p-1\). If the period of the solution has been divided into r ranges, at the end we get an integer vector \((s_1,\ldots ,s_r)\) with \(s_j\in \{0,1,\ldots ,2^p-1\}\) for \(j=1,\ldots ,r\) which is the pattern of the solution for the chosen initial conditions.
For each of the considered networks we finally obtain (at most) 200 patterns, but some of them could be equivalent. Two patterns are considered equivalent if they are related by: (i) a reordering of the neurons of the network; (ii) a cyclic translation of the pattern coordinates; (iii) an automorphism of the network graph; or (iv) a combination of the previous operations. After a final post-processing to check equivalences, the pattern list is divided into equivalence classes. The final output of the processing is, for each network, a list of patterns (representatives of each equivalence class), together with the size of the equivalence class of each of them.
2.4 Color synchronization patterns: definitions
A graph is q-colorable if its vertices can be grouped into q disjoint non-empty subsets in such a way that adjacent vertices lie on different subsets of the partition. If this is the case, such decomposition is a q-coloring of the graph and the subsets are the colors. A graph is bipartite if and only if it is 2-colorable. Two q-colorings of a graph are equivalent if they are related by an automorphism of the graph, and they are said to be different otherwise. It is easy to deduce that all 2-colorings of a bipartite graph are equivalent.
On the other hand, a synchronization pattern on a neuron network defines in a natural way a partition of the set of neurons: two neurons \(x,x'\) are in the same subset of the partition if they have overlapping bursting intervals or if there is a sequence of neurons
from x to \(x'\) such that each \(x_i\) has an overlapping bursting interval with \(x_{i+1}\). We say that the pattern is a q-color pattern if the partition defined by the pattern is a q-coloring of its underlying graph. The pattern is a color pattern if it is a q-color pattern for some q. Following the notation from graph theory, a 2-color pattern is also called a bipartite pattern. With this notation, the tripod gait on the GH network is a bipartite pattern while the tetrapod gait is a 3-color pattern. All the patterns that were obtained in [16, 17] for the GH network are q-color patterns for \(q=2,3\) or 4.
Remark 1
As already commented, since synapses are inhibitory, it is natural to expect that connected neurons would not be active at the same time. Therefore, it is also natural to expect that color patterns form an important subfamily of synchronization patterns.
3 Results
In order to study all the possible patterns, we have applied the quasi-Monte Carlo sweep (Sect. 2.3) to all the 982 bipartite bidirectional networks constructed as in Sect. 2.2 from the bipartite graphs with up to 9 vertices of the dataset of [25]. The following results were obtained taking \(t_\text {small}=0.25\). We took this high value to simplify the resulting patterns as much as possible. Nevertheless, with smaller values of this parameter the results were quite similar, with the exception that is discussed in Sect. 3.3.
All observed patterns in a 9-neurons CPG considering a set of 200 initial conditions in a Halton sequence. The total percentage of each pattern is indicated. The time series of the 9 neurons are presented for each pattern, and they show the bursting dynamics of each neuron and the synchronization patterns. We have colored the groups of neurons according to their synchronization with \(t_\text {small} = 0.25\)
We show possible examples of results on a particular network in Fig. 3 to illustrate the existence of numerous patterns on each network. For display purposes, we only show the connectivity of the network, but all synapses are bidirectional (see Sect. 2.2). We also show the corresponding time series and synchronization patterns. Synchronization patterns are usually represented schematically in diagrams, like those in Figs. 1d and 2d, where each horizontal line represents the state (active or inactive) of a single neuron. These diagrams fit well with the examples presented.
Note that any network can have a large number of patterns (multistability), but the size of its basin of attraction can be very small. We estimate it by considering the number of initial conditions that converge to each one and giving the corresponding percentage. In Fig. 3 we show for a particular 9-neurons CPG example all the patterns (5 patterns) found considering a set of 200 initial conditions in a Halton sequence. By increasing this number more patterns may appear, but with a low percentage value. Furthermore, most of the patterns maintain the same bursting dynamics (same number of spikes per burst), but some of them change the number of spikes per burst in some neurons (see neuron 6 in the last pattern). We remark that we have set along the article a fixed threshold percentage (5\(\%\)) to perform the analysis, but in this example we show all the found patterns. We have colored the groups of neurons according its synchronization with \(t_\text {small} = 0.25\). Note that increasing the value \(t_\text {small}\) allows more patterns to be considered equal and therefore increase their overall percentage and pass the threshold percentage. For the fourth pattern we show two sets of time series with a slight difference on one of its neurons that are considered equal with \(t_\text {small} = 0.25\). This difference is seen taking \(t_\text {small} = 0.05\).
In the following, we will represent synchronization patterns for generic networks together with network topology with a 3D representation (Fig. 5 and following) where we draw some copies of the network in parallel horizontal planes and vertical cylinders represent the activation intervals of the corresponding neurons. We remark that the use of graph techniques in networks has also been recently considered in [26].
3.1 Dominance of bipartite patterns
The first result that we discuss is that for the selected parameter values, which are within the tripod gait dominance region for the insect movement network, there is an evident predominance of bipartite patterns, that is, tripod-type patterns. The results we have obtained are the following:
-
100/70 rule: in 100\(\%\) of the graphs considered, the bipartite pattern appeared in more than 70\(\%\) of the sampled initial conditions, and in near 70\(\%\) of the graphs (627 of the 982 graphs) the bipartite pattern appeared in 100\(\%\) of the sampled initial conditions.
-
95/90 rule: for around 95\(\%\) of the graphs (929 out of 982), the bipartite pattern appeared in at least 90\(\%\) of the sampled initial conditions, and for nearly 90\(\%\) of the graphs (863 out of 982), the bipartite pattern appeared in at least 95\(\%\) of the sampled initial conditions.
Moreover, in more than 99\(\%\) of the graphs (978 out of 982), the bipartite pattern appeared in at least 80\(\%\) of the simulations. Thus, it is clear that the bipartite pattern is predominant for all the networks, at least with the chosen set of parameters.
Remark 2
The actual patterns depend heavily on the initial conditions used for the quasi-Monte Carlo method. Different initial conditions may report slightly different patterns, since we depend on 200 sampled points on high dimensional spaces. However, the global picture is the same for other sets of initial conditions.
3.2 Non-bipartite patterns
All the graphs having a non-bipartite pattern from 6-neurons up to 9-neurons CPG networks. In the sake of visualization, we just show the connectivity of the network, but all the synapses are bidirectional, see Sect. 2.2
In our simulations there appeared other synchronization patterns different from the bipartite ones. We will focus in those non-bipartite patterns that have a relevant statistical significance: we will take into account only those patterns that appear in more than 5\(\%\) of the simulations of the corresponding network. Among all the considered networks, 83 have a non-bipartite pattern (Fig. 4): 71 of them have just one non-bipartite pattern and 12 of them have two non-bipartite patterns. This makes a total number of 95 non-bipartite patterns, 87 of which are 3-color patterns. The remaining 8 non-bipartite patterns are not color patterns and they can be divided into two classes. On one hand, there are three patterns where there are no connected neurons firing at the same time but such that the sequences of partial overlappings of the bursting intervals produce that connected neurons are in the same subset of the partition defined by the pattern. These examples are depicted in Fig. 5, where it can be seen that in fact all the neurons of the network are connected by the sequence of bursting interval overlappings and thus the partition defined by the pattern is the trivial partition containing only the whole set of neurons. On the other hand, there are 5 patterns (Fig. 6) where the partition defined by the pattern contains 3 subsets, but such that there are “bad edges” connecting neurons that fire at the same time despite inhibitory coupling. Those patterns are the only ones that contradict Remark 1, and they deserve a further investigation.
Three weak patterns with \(t_\text {small}=0.25\). Although connected neurons never fire at the same time, the concatenation of partial overlappings of the bursting intervals produce that connected neurons are in the same subset of the partition defined by the pattern. There is another weak pattern but it has a bad edge (Fig. 6e)
3.3 \(t_\text {small}\) and strong vs weak synchronization patterns
For a given activity pattern, we can distinguish two levels of synchronization according to the type of intersections between the bursting intervals of different neurons that can be found. On the one hand, there are patterns where the bursting intervals of the neurons are fully synchronized: if two neurons are both active at some point, the coincidence of their bursting intervals is complete. On the other hand, there are patterns where partial overlappings between bursting intervals occur: there are neurons which are all active at some common time but whose bursting intervals do not coincide (Fig. 5). We will call strong patterns to the former ones and weak patterns to the latter.
We have investigated if the obtained patterns are strong or weak synchronization patterns. For non-bipartite patterns our findings depend heavily on the pre-processing of the pattern data. As we have explained in Sect. 2.3, in order to identify similar patterns, in some step we discard parts of the dynamical pattern which happens for a small amount of time, quantified by a fixed parameter \(t_\text {small}\in (0,1)\). When this parameter is small, very few non-bipartite patterns are strong synchronization patterns. As this parameter grows, more synchronization patterns are associated (as we are discarding short length differences between the patterns) and more of them become strong. For example, if \(t_\text {small}\) is set equal to 0.05 only 3 non-bipartite patterns are strong patterns (Fig. 7). On the contrary, when \(t_\text {small}\) is set equal to 0.25, there are only 4 weak synchronization patterns. Three of them can be seen in Fig. 5, the remaining one has a “bad edge” (Fig. 6e). On Fig. 6a-d we present the 4 non-bipartite strong with a “bad edge”, that is, with an edge connecting neurons that fire at the same time despite inhibitory coupling.
It is important to remark that this dependence on the parameter \(t_\text {small}\) does not occur for bipartite patterns: bipartite patterns appear always as strong patterns even for very small values of \(t_\text {small}\).
3.4 Some remarkable findings
We end this section with some remarkable results about patterns on cyclic graphs and non-bipartite graphs.
3.4.1 The octagon: starry traveling waves on cyclic graphs
The diagram of Fig. 5a shows a special pattern on the cyclic graph of 8 vertices (the octagon). In this pattern the neurons are firing consecutively following a starry-like traveling wave. There is an ordering of the eight neurons
such that \(x_i\) fires exactly after \(x_{i-1}\) for \(i=2,\dots ,8\) (and \(x_1\) fires again after \(x_8\)) with a certain delay, causing an overlapping of their bursting intervals. The ordering of the neurons is the one used for drawing the 8-pointed star out from the vertices of the regular octagon (Fig. 8a): (i) starting from any vertex we jump to the vertex located 3 positions further (clockwise or counterclockwise, clockwise in Fig. 8a); (ii) we repeat this movement with all the jumps in the same sense (clockwise or counterclockwise) until we are back at the starting vertex. As we can see in Fig. 8b, the pattern period is divided into 16 ranges such that at each range there are alternatively 2 or 3 firing neurons.
In our experiment for the octagon, the bipartite pattern was present in 82% of the simulations, while the starry traveling wave pattern (considering as the same the clockwise and counterclockwise cases) appeared in the remaining simulations. We have investigated if similar starry traveling wave patterns have the same importance for other cyclic graphs. From now on we will use the terms p-gon or \(C_p\) to denote the cyclic graph with p vertices. In our original experiment there only appeared the cyclic graphs of orders 4, 6 and 8 (the only bipartite cyclic graphs with up to 9 vertices). The bipartite pattern appeared in around 100\(\%\) and 90\(\%\) of the simulations for the 4-gon and for the 6-gon, respectively. In the remaining simulations for the 6-gon there appeared the 3-color pattern of Fig. 7a.
There is a well-known general procedure for drawing a p-pointed star out from the vertices of \(C_p\):
-
(i)
choose a positive integer number q coprime with p and with \(q< p/2\);
-
(ii)
starting from any vertex \(x_1\) of \(C_p\), jump to the vertex located q positions further (say clockwise) and join both vertices with a segment;
-
(iii)
repeat (ii), each time starting from the final vertex of the previous step, until reaching \(x_1\) again. If we call (p, q)–star to the resulting figure, we can consider an associated (p, q)–star traveling wave pattern in the network with underlying graph \(C_p\) in the same way as we explained for \(C_8\) before. With this notation, the pattern in \(C_8\) considered before is an (8, 3)–star traveling wave pattern.
We made the simulations also for the cyclic graphs of order 5, 7, 9, 10, 11 and 12, with the following findings:
-
For \(p=5,7,11\) (prime numbers) the whole set of found patterns were star traveling wave patterns. For \(p=5,7\) almost 100\(\%\) of the patterns are the (5, 2)– and (7, 2)–star traveling wave patterns, respectively. For \(p=11\), the (11, 2)–star traveling wave pattern appears in 92\(\%\) of the simulations while the (11, 3)–star traveling wave pattern appears in the remaining ones.
-
For \(p=9\), the (9, 2)–star traveling wave pattern appears in 97\(\%\) of the simulations.
-
For even p (\(p=10,12\)) the resulting graph is bipartite and as it could be expected the bipartite pattern is again dominant, appearing in 71\(\%\) and 65% of the simulations respectively. For \(p=12\), almost all the remaining cases took the form of the (12, 5)–star traveling wave pattern.
-
For p multiple of 3 (\(p=9,12\)) the 3-color patterns of Fig. 9 appear but without statistical significance: only in 1.5\(\%\) and 0.5\(\%\) of the simulations for \(C_9\) and \(C_{12}\) respectively.
-
For \(p=10\), there appears no star traveling wave pattern. Instead, the non-bipartite cases converged to the pattern depicted in Fig. 10a. In this pattern, each neuron is fully synchronized with its opposite one. Starting from a couple of firing neurons \(x_1,\bar{x}_1\) located at opposite vertices of \(C_{10}\), with a delay of 2/3 of their bursting interval, the neurons \(x_2,\bar{x}_2\) located 2 positions further (clockwise in the picture) from \(x_1,\bar{x}_1\) respectively start their activity intervals, and so on. This pattern can be seen as the coupling of two interlaced/synchronized (10, 2)–star traveling wave patterns, having in mind that a (10, 2)–star is not even a star but a 5-gon, indeed. With this viewpoint, the 3-color pattern of the 6-gon depicted in Fig. 7a can be considered also as the coupling of two (6, 2)–star traveling wave patterns.
Therefore, it seems that, at least for the chosen set of parameters, for odd p (non-bipartite connectivities) the (p, 2)–star pattern is the dominant pattern in \(C_p\). For even p the bipartite pattern is dominant in \(C_p\) but it is not clear which must be the second pattern in importance for a generic p. It is also interesting to note that when p is a multiple of 3 (\(p=9,12\)) the quite symmetric patterns of Fig. 9 rarely appear.
If p is even, as p increases, the percentage of simulations giving rise to the bipartite pattern decreases in favor of the star traveling wave patterns.
3.4.2 Non-bipartite examples
We wonder if it is possible to predict the dominant patterns of non-bipartite networks from the network topology. In Sect. 3.4.1 we have seen a few examples (cyclic networks with odd number of neurons), where the starry traveling wave patterns appeared as dominant. We have studied a couple more examples: the 3-color networks whose underlying graphs are the complete 3-color graphs \(K_{222}\) and \(K_{333}\) of Fig. 11. In general, the complete 3-color graph \(K_{abc}\), with \(a,b,c\in \mathbb {N}\), is constructed by taking three sets with a, b and c vertices and connecting each vertex of a set with all the vertices of the other two sets (and not connecting vertices within the same set). These graphs are interesting because they are not bipartite, their 3-coloring is unique, and every 3-colored graph with a, b and c vertices in its colors is a subgraph of \(K_{abc}\).
We have performed the simulation for those two networks. Their 3-color patterns of Fig. 11c, d are the most relevant ones, but their dominance is weaker than that of the bipartite pattern in bipartite networks. The 3-color pattern appears in around 50\(\%\) of the simulations for \(K_{222}\), and in 40\(\%\) for \(K_{333}\). Thus, the dominance of bipartite patterns for bipartite networks cannot be extended to 3-color patterns in 3-colorable, non-bipartite, networks.
4 Conclusions
This article shows how in some situations the topology of the network has a strong influence on its dynamics. The tripod and tetrapod gait pattern appearing in the Ghigliazza and Holmes CPG model for the movement of insects have natural generalizations in color patterns for general directed networks.
In particular, tripod gait has a natural generalization as bipartite patterns for directed bipartite networks. We have generalized GH network to networks over a general graph, and using a set of parameters for which the tripod gait is ubiquitous for this model we have checked if bipartite patterns are also ubiquitous in other cases. We have explored all bipartite networks of up to 9 neurons, and the bipartite pattern appears as the dominant pattern in all cases. In the particular case of cyclic graphs, we have studied the possibilities of the graphs of order 5, 7, 9, 10, 11 and 12 and we have seen that for odd p the (p, 2)–star pattern is the dominant pattern in \(C_p\). This result is a first attempt to study larger networks where these structures have a lot of sense (like in the movement of centipedes).
This dominance of bipartite patterns and other interesting cases that appear too, suggest some topics that deserve further research, such as:
-
the dominance of bipartite patterns for longer networks and other regions of the parameter space;
-
patterns with bad edges, where connected neurons are fully synchronized;
-
patterns in cyclic networks, with special attention to starry traveling wave patterns;
-
the importance of 3-color patterns in 3-colorable, non-bipartite networks; and
-
which dynamical patterns are stable under random perturbation of the parameters?
Data availability
Graphs, initial conditions and resulting patterns are available as Supplementary Material.
References
da Fontoura Costa, L.: Discovering patterns in bipartite networks. BioRxiv (2022). https://doi.org/10.1101/2022.07.16.500294
Kitsak, M., Papadopoulos, F., Krioukov, D.: Latent geometry of bipartite networks. Phys. Rev. E 95(032), 309 (2017)
Pavlopoulos, G.A., Kontou, P.I., Pavlopoulou, A., Bouyioukos, C., Markou, E., Bagos, P.G.: Bipartite graphs in systems biology and medicine: a survey of methods and applications. GigaScience 7(4), giy014 (2018)
Pastor, J.M., Santamaría, S., Méndez, M., Galeano, J.: Effects of topology on robustness in ecological bipartite networks. Netw. Heterogeneous Media 7(3), 429–440 (2012). https://doi.org/10.3934/nhm.2012.7.429
Dunne, J.A., Williams, R.J., Martinez, N.D.: Food-web structure and network theory: the role of connectance and size. Proc. Natl. Acad. Sci. U.S.A. 99, 12917–12922 (2002)
Goh, K.I., Choi, I.G.: Exploring the human diseasome: the human disease network. Brief. Funct. Genom. 11(6), 533–542 (2012)
Lamb, D.G., Calabrese, R.L.: Small is beautiful: models of small neuronal networks. Curr. Opin. Neurobiol. 22(4), 670–675 (2012). https://doi.org/10.1016/j.conb.2012.01.010
Bal, T., Nagy, F., Moulins, M.: The pyloric central pattern generator in crustacea: a set of conditional neural oscillators. J. Comp. Physiol. A 163, 715–727 (1996)
Marder, E., Calabrese, R.: Principles of rhythmic motor pattern generation. Physiol. Rev. 76, 687–717 (1996)
Marder, E., Bucher, D.: Central pattern generators and the control of rhythmic movements. Curr. Biol. 11(23), R986–R996 (2001)
Rybak, I.A., Dougherty, K.J., Shevtsova, N.A.: Organization of the mammalian locomotor CPG: review of computational model and circuit architectures based on genetically identified spinal interneurons. eNeuro (2015). https://doi.org/10.1523/ENEURO.0069-15.2015
Bidaye, S.S., Bockemühl, T., Büschges, A.: Six-legged walking in insects: how CPGS, peripheral feedback, and descending signals generate coordinated and adaptive motor rhythms. J. Neurophysiol. 119(2), 459–475 (2018). https://doi.org/10.1152/jn.00658.2017
Ashwin, P.: Symmetric chaos in systems of three and four forced oscillators. Nonlinearity 3(3), 603–617 (1990)
Ashwin, P., Burylko, O., Maistrenko, Y.: Bifurcation to heteroclinic cycles and sensitivity in three and four coupled phase oscillators. Physica D 237(4), 454–466 (2008)
Rosenblum, M.G., Pikovsky, A.S., Kurths, J.: Phase synchronization in driven and coupled chaotic oscillators. IEEE Trans. Circuits Syst. I Fund. Theory Appl. 44(10), 874–881 (1997)
Barrio, R., Lozano, Á., Rodríguez, M., Serrano, S.: Numerical detection of patterns in CPGs: gait patterns in insect movement. Commun. Nonlinear Sci. Numer. Simul. 82(105), 047 (2020)
Barrio, R., Lozano, Á., Martínez, M., Rodríguez, M., Serrano, S.: Routes to tripod gait movement in hexapods. Neurocomputing 461, 679–695 (2021)
Ghigliazza, R., Holmes, P.: A minimal model of a central pattern generator and motoneurons for insect locomotion. SIAM J. Appl. Dyn. Syst. 3(4), 671–700 (2004)
Chun, C., Biswas, T., Bhandawat, V.: Drosophila uses a tripod gait across all walking speeds, and the geometry of the tripod is important for speed control. eLife 10, e65878 (2021)
Ramdya, P., Thandiackal, R., Cherney, R., Asselborn, T., Benton, R., Ijspeert, A., Floreano, D.: Climbing favours the tripod gait over alternative faster insect gaits. Nat. Commun. 8(14), 494 (2017)
Ghigliazza, R.M., Holmes, P.: Minimal models of bursting neurons: how multiple currents, conductances, and timescales affect bifurcation diagrams. SIAM J. Appl. Dyn. Syst. 3(4), 636–670 (2004). https://doi.org/10.1137/030602307
Halton, J.H.: Algorithm 247: radical-inverse quasi-random point sequence. Commun. ACM 7(12), 701–702 (1964)
Hairer, E., Nørsett, S., Wanner, G.: Solving Ordinary Differential Equations I Nonstiff problems, 2nd edn. Springer, Berlin (2000)
Prince, P., Dormand, J.: High order embedded Runge–Kutta formulae. J. Comput. Appl. Math. 7(1), 67–75 (1981)
Alcalde Cuesta, F., González Sequeiros, P., Lozano Rojo, A., Vigara Benito, R.: An accurate database of the fixation probabilities for all undirected graphs of order 10 or less. In: Rojas, I., Ortuño, F. (eds.) Bioinformatics and Biomedical Engineering, pp. 209–220. Springer, Berlin (2017)
Curto, C., Morrison, K.: Graph rules for recurrent neural network dynamics. Not. Am. Math. Soc. 70(4), 536–551 (2023)
Funding
Open Access funding provided thanks to the CRUE-CSIC agreement with Springer Nature. AL, RV, CMC, and RB have been supported by the Spanish Research project PID2021-122961NB-I00. AL has been supported European Regional Development Fund and Diputación General de Aragón (ER22_23R) RV, CMC and RB have been by the European Regional Development Fund and Diputación General de Aragón (E24-23R). RB has been supported by the Spanish Research project TED2021-130459B-I00 and by the European Regional Development Fund and Diputación General de Aragón (LMP94-21).
Author information
Authors and Affiliations
Contributions
All authors have contributed equally to this manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no Conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Lozano, Á., Vigara, R., Mayora-Cebollero, C. et al. Dominant patterns in small directed bipartite networks: ubiquitous generalized tripod gait. Nonlinear Dyn 112, 15549–15565 (2024). https://doi.org/10.1007/s11071-024-09830-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11071-024-09830-2