Abstract
Some types of tumors in people with brain cancer grow so rapidly that their average size doubles in twenty-five days. Precisely determining the type of tumor enables physicians to conduct clinical planning and estimate dosage. However, accurate classification remains a challenging task due to the variable shape, size, and location of the tumors.The major objective of this paper is to detect and classify brain tumors. This paper introduces an effective Convolution Extreme Gradient Boosting model based on enhanced Salp Swarm Optimization (CEXGB-ESSO) for detecting brain tumors, and their types. Initially, the MRI image is fed to bilateral filtering for the purpose of noise removal. Then, the de-noised image is fed to the CEXGB model, where Extreme Gradient Boosting (EXGB) is used, replacing a fully connected layer of CNN to detect and classify brain tumors. It consists of numerous stacked convolutional neural networks (CNN) for efficient automatic learning of features, which avoids overfitting and time-consuming processes. Then, the tumor type is predicted using the EXGB in the last layer, where there is no need to bring the weight values from the fully connected layer. Enhanced Salp Swarm Optimization (ESSO) is utilized to find the optimal hyperparameters of EXGB, which enhance convergence speed and accuracy. Our proposed CEXGB-ESSO model gives high performance in terms of accuracy (99), sensitivity (97.52), precision (98.2), and specificity (97.7).Also, the convergence analysis reveals the efficient optimization process of ESSO, obtaining optimal hyperparameter values around iteration 25. Furthermore, the classification results showcase the CEXGB-ESSO model’s capability to accurately detect and classify brain tumors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Brain tumor is one of the most threatening diseases in the world, affecting people of all genders, ages, and ethnicities [1]. A brain tumor is a growth of abnormal brain tissue that can be malignant or benign [2, 3]. The ability of neurologists to accurately diagnose tumors depends on their knowledge and skill [4]. Nowadays there is a significant rise in brain tumor patients, which leads researchers to automate brain tumor diagnosis. Following are some of the approaches that a computer-aided diagnosis (CAD) system can assist neurologists [5]. This reduces the need for human interaction and promotes early discovery of cancers for better treatment [6]. It provides the physician with a secondary perspective when making the ultimate decision. A brain tumor affects approximately 700,000 people worldwide, with nearly 86,000 new diagnoses identified in 2019 [7].
There are approximately 120 types of brain tumors, including meningioma (35%), pituitary tumors (14%), and glioma (16%) [8]. These abnormalities are defined by their shape, size, and location of tumor [9]. Both Pituitary and Meningioma tumors grow around the pituitary gland and the skull region, respectively. Glioma tumors are of diverse intensities and spread across the glial cells of the brain [10]. A human expert can use visual check of these properties to recognize the type of tumor. A proper classification can help you make the best decision and get the best treatment. Understanding this classification is a difficult and time-consuming task for medical experts. At difficult stages, these activities are subjected to traditional methods such as human inspection, biopsy, expert opinion, and so on, all of which take time. As a result, CAD is needed for early detection of brain tumor in less time without the need of humans. Computer Tomography (CT) and Magnetic Resonance Imaging (MRI) are two modalities commonly used to identify abnormalities in the size, location, or shape of brain tissues that can help identify tumors. But MRI produces more detailed images than CT scans, so physicians prefer MRI [11].
Different methods like artificial neural network (ANN) [11], CNN [12], support vector machine (SVM) [14], and K-nearest neighbors (KNN)[13] are used by researchers. When dealing with large amounts of data, certain problems like vanishing gradient, and over fitting can lead to errors. This results in misclassification of tumor types. This motivates us to develop an efficient method for brain tumor detection.
The contributions of the proposed CEXGB-ESSO model are as follows;
-
The CEXGB-ESSO model adopts a hybrid architecture, integrating CNN and EXGB. This fusion capitalizes on the strengths of CNN for feature extraction and EXGB for robust classification, presenting a unique combination that effectively addresses the challenges posed by brain tumor variability.
-
The Convolutional neural network replaces the fully connected layer with EXGB, which minimizes the number of parameters to detect the tumor boundary. The CEXGB model doesn’t need to fetch weight values from a fully connected layer to adjust the weights in previous layers, which introduces simplicity in deep detection of the tumor region.
-
ESSO is used to find the optimal EXGB hyperparameters that enhance the convergence speed. Both the ability to explore and exploit are enhanced by the introduction of an inertia weight parameter by ESSO, leading to faster convergence towards optimal hyperparameter values.
-
The proposed CEXGB-ESSO model performs better in terms of accuracy, sensitivity, precision, and specificity when compared to existing methods.
The structure of the paper is described below: Sect. 2 gives an overview of the existing works. Section 3 explains our proposed CEXGB-ESSO model. The proposed approach is evaluated to an experiment evaluation in Sect. 4. Section 5 concludes the paper.
2 Related Works
This section provides a concise overview of studies undertaken to identify brain tumorsutilizing various existing methods.
Kumar et al. [15] presented a deep network model using global average pooling and ResNet-50 (GResNet) for tumor classification. ResNet-50 with transfer learning is used to classify brain tumors. The output layer of ResNet-50 incorporates stacked average-pooling and fully connected softmax layers designed for classifying images into a thousand categories in the ImageNet dataset. Stacking of layers leads to complexity in tumor detection. Here, the accuracy of tumor region was not accurately identified. Majib et al. [16] introduced a VGG Stacked Classifier (VSC) network for brain tumor classification from MRI images. For transfer learning models, the initial weights of the pre-trained model are created and used to initialize the CNN model. Thus, a restricted number of epochs may be used to efficiently train the model. This procedure is more complex due to high-resolution, unsteady training. Dixit & Nanda [17] presented a radial-basis neural network based on improved whale optimization (RNIWO) for brain tumor classification. First, the input is given to the preprocessing steps. Then, segmentation is performed to recognize the tumor region by using fuzzy-c means (FCM) clustering. This clustering based approach misdiagnoses tumor size, putting human lives at risk. Ahuja et al. [18] introduced DarkNet methods for classifying brain tumors. DarkNets approaches are trained using enhanced data and compared using various evaluation metrics. Tumor localization is performed utilizing feature maps from better-performing DarkNet approaches. The volume of the tumor was not accurately identified in this approach. Sharif et al. [19] introduced Densenet model for brain tumors classification. This model utilized a deep transfer to learn unbalanced data to implement the proposed strategy. Each type of tumor information is contained in the pooling layer, which is used to extract the characteristics of the trained model. The main disadvantage of this approach is the reduction of significant features. Sasank et al. [20] introduced an improved kernel-based softplus learning machine (KELM) classification method for brain tumor classification. This method gives better segmentation results. But the classifier fails to perform on large datasets. Alhassan et al. [21] proposed a swish-based RELU activation (SRELU) for brain tumor type classification. The performance is better, but the significant features are not selected.
Khairandish et al. [23] introduced a novel hybrid CNN-SVM model for precise brain tumor classification. Their approach begins with preprocessing and normalization, ensuring input data consistency and standardization, leading to improved overall model performance. Leveraging Maximally Stable Extremal Regions (MSER) for feature extraction, the method demonstrates robustness to noise and illumination changes in medical images. The hybrid CNN-SVM classification step combines the strengths of CNN’s feature learning capabilities and SVM’s effectiveness in handling high-dimensional data, resulting in accurate tumor classification. Noteworthy advantages include enhanced feature extraction, robustness to noise, effective region identification, and leveraging the strengths of both CNN and SVM. However, the model’s complexity of implementation and potential computational resource requirements pose challenges, requiring careful design and optimization.
Sadad et al. [24] proposed the NASNet for brain tumor classification, employing an optimized framework that integrates Evolutionary Algorithms (EAs) and Reinforcement Learning (RL) to enhance the model’s performance. The incorporation of EAs and RL in the NASNet model provides an optimized framework for brain tumor classification, contributing to improved performance. NASNet’s capability to extract significant features from MRI slices enhances its effectiveness in accurately classifying different types of brain tumors. Despite these advantages, the NASNet model presents a notable disadvantage in its large number of parameters. This parameter abundance may lead to increased computational complexity and resource requirements.
Nawaz et al. [25] focused on optimizing brain tumor diagnosis through the development of the hybrid-brain-tumor-classification (HBTC) framework, streamlining the process and enhancing overall diagnostic performance. The framework incorporates preprocessing and segmentation of brain MRI datasets, utilizing features like Co-occurrence matrix (COM), run-length matrix (RLM), and gradient features. Although the study successfully identifies nine optimized features for tumor classification, it lacks detailed performance metrics and insights into implementation challenges. In contrast, Kesav and Jibukumar [26] aimed to minimize the execution time of a conventional RCNN architecture by employing a Two Channel CNN for accurate classification of Glioma and healthy tumor MRI samples. This approach, extending to detect and bound tumor regions in various tumor types, demonstrates a significant reduction in execution time compared to existing architectures.
The existing approaches for brain tumor classification exhibit several notable research gaps, including incomplete identification of the tumor region, challenges in accurate estimation of tumor size, instability during training processes, reduction of essential features, and limitations in handling large datasets. These gaps collectively contribute to suboptimal performance, misdiagnoses, and compromised robustness. Our proposed CEXGB-ESSO model addresses these research gaps by introducing a streamlined architecture that replaces the fully connected layer with EXGB, minimizing parameters and enhancing feature extraction. The incorporation of ESSO fine-tunes EXGB hyperparameters, mitigating training instabilities and contributing to improved model accuracy. The removal of the fully connected layer ensures accurate tumor boundary detection, and the optimized hyperparameter selection by ESSO resolves challenges related to inaccurate tumor size estimation. Additionally, the proposed model demonstrates robust performance on large datasets, overcoming limitations observed in existing approaches. Collectively, these novel features position the CEXGB-ESSO model as a promising solution to the identified research gaps, offering enhanced accuracy, efficiency, and scalability in brain tumor classification.
3 Proposed CEXGB-ESSO Model for Brain Tumor Detection and Classification
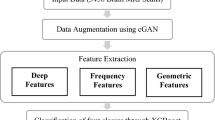
We proposed a CEXGB-ESSO model for brain tumor detection and classification (No tumor, Pituitary, glioma, and Meningioma). The proposed system consists of two phases: Preprocessing and classification. Figure 1 represents the architecture of the CEXGB-ESSO model. Initially, the MRI image is fed to a preprocessing phase, where it is filtered using bilateral filtering. In the classification phase, the proposed CEXGB-ESSO model is utilized to detect and classify the tumors. CNN is utilized to extract tumor features (shape, location, and size), and EXGB is used to classify tumor type. ESSO is used to fine-tune EXGB hyperparameters. Hence, the detection accuracy has improved.
3.1 Preprocessing
In the preprocessing phase, bilateral filtering is applied to enhance the input brain MRI image. This non-linear, noise-reducing, and edge-preserving smoothing filter computes the intensity values of the weighted average of surrounding pixels to replace each pixel’s intensity. The weights are determined using a Gaussian distribution, accounting for both Euclidean distance between pixels \(\left\| {u - v} \right\|\) and radiometric differences (depth distance, color intensity, etc.). This process effectively preserves the sharp edges. The bilateral filter is mathematically represented by Eq. (1), which symbolizes the normalized weighted average.
where \(W_{u}\) denotes the normalization factor determined in Eq. (2).
where \(g_{s}\) represents spatial Gaussian, \(g_{r}\) denotes the range Gaussian, \(I_{u}\) represents intensity value of pixel u,s and r parameters denote the filtration amount, and \(I_{v}\) represents intensity value of pixel v.
3.2 Classification of Brain Tumors
The preprocessed image is fed to the CEXGB-ESSO model to detect and classify the tumor. Existing methods like ANN, CNN, SVM, and KNN have some problems like overfitting, being time-consuming, increasing the number of parameters, and failing to increase the performance of brain tumor detection and classification. To overcome this issue, a CEXGB-ESSO model is introduced. The CEXGB model can accurately detect the tumor boundary and tumor region. A fusion between CNN and XGBoost is known as CEXGB. The architecture of CEXGB contains numerous stacked layers of convolution, with the final layer being EXGB. There is no fully connected layer because there is no need to re-adjust the weights in previous layers. This simplifies the process and minimizes the number of calculation parameters. ESSO can be used to fine-tune the XGB parameters, which enhance convergence speed and accuracy.
3.2.1 Architecture of CEXGB
The CEXGB consists of a convolution layer, a pooling layer, and an EXGB layer. The first layer of CEXGB is a convolutional layer that uses a kernel to extract features (shape, location, and size) from an MRI image and outputs a feature map. A convolution operation consists of numerous components, such as the input image, feature map, and kernel. After extracting the features, the irrelevant data is omitted, and the dimensionality of the data is reduced by using pooling layers without losing significant data. Max-pooling exhibits excellent performance and is commonly used to reduce dimensionality. Max-pooling selects the highest value from the matrix and ignores the remaining values. The max-pooling mathematical formulation is given in Eq. (3).
where y signifies the previous layer output, i, jdenote the spatial position, \(Max_{s} ()\) signifies the max function applied to the max-pooling window of dimension. The fully connected layer of CNN is removed by introducing EXGB as the last layer. Significant features of max-pooling are fed to the EXGB layer. EXGB is a classifier that turns weak learners into strong ones. A classifier that does not give correct detection is referred to as a weak learner. A classifier integrates weak learners to achieve accurate classification, so CNN utilizes EXGB to classify brain tumors. Weak learners are built to train the input images using extracted features and then combine to form a strong one. The leaf node decides whether the tumor is normal or abnormal. The leaf node, also known as the terminal node of the tree, contains the class labels.Consider a database ‘A’,\(A = \left\{ {(u_{i} ,v_{i} )} \right\}(\left| A \right| = n,u_{i} \in R^{p} ,v_{i} \in R^{n} )\). Here, \(u_{i}\) signifies the members of training set, \(v_{i}\) denotes the appropriate class labels, ‘p’denotes the number of features and nsignifies the number of the samples.The output \(\hat{v}_{l}\) of a tree boosting model with K trees is stated in Eq. (4).
where \(f_{k}\) signifies the \(k^{th}\) tree’s leaf score, F is the classification tree. The tree model’s set of function \(f_{k}\) can be learned by reducing the objective function, which is given in Eq. (5).
where L signifies the loss function in the training, which calculates the distance between target \(\hat{v}_{l}\) and object \(v_{i}\). The term \(\Omega\) denotes the complexity of tree model’s penalty term, which is determined in Eq. (6).
where \(\lambda\) and \(\gamma\) are constants that regulate the degree of regularization, N denotes the number of leaves, and D signifies the leaf weight. Gradient boosting is a technique that can be used to solve classification problems. It is utilized with the loss function; the loss function is expanded via a second-order Taylor expansion with the removal of the constant, which results in the following simplified objective given in Eq. (7).
where \(E_{g} = \left\{ {\frac{i}{{q(u_{i} )}}} \right\} = j\) signifies the instance set of ‘g’ leaf, and \(q(u_{i} )\) is the map assigning the sample to appropriate leaf.\(x_{i}\) and \(y_{i}\) are the gradient statistics of first and second order on the loss function, which is computed in Eq. (8) and (9).
The optimal weight \(D_{g}^{*}\) of ‘g’ leaf is determined using Eq. (10).
Then substituting \(D_{g}^{*}\) into Eq. (7), there exists,
In practice, split candidates are evaluated using instance set’s right nodes \(E_{R}\) and left nodes \(E_{L}\) score. After the split, the loss reduction is given in Eq. (12).
where \(E = E_{R} \cup E_{L}\), \(E_{R}\) denotes the instance sets of right nodes, and \(E_{L}\) signifies the instance sets of left nodes.
3.2.2 Enriched Salp Swarm Optimization
The ESSO is utilized to find the optimal EXGB hyperparameter values (learning rate, lambda, maximum depth, and subsample), and a better classifier model is developed for brain tumor detection and classification. Salps belong to the Salpidae family and have a translucent, barrel-shaped body. Their tissues are similar to those of jellyfish. Like jellyfish, they migrate by pushing water through their bodies as propulsion. The swarming habit of salps is characterized by the formation of a swarm called a salp chain (SC) in the deep oceans. The behavior of the swarm is used to discover the best EXGB hyperparameter. There are two categories of salps in this swarm: the follower salp (FS) and the leader salp (LS). The FS accompanies the leader in search of food through the exploration and exploitation phases (optimal EXGB hyperparameters). The fitness of each solution is determined, and the solution with optimal fitness is utilized as the EXGB hyperparameter.
Equation (13) is used to update the position of the LS.
where \(X_{a}^{1}\) and \(Q_{a}\) signifies the location of leader in ‘a’ dimension and food source, \(v_{a}\) and \(h_{a}\) signifies the lower bound and upper bound of ath dimension, \(p_{1}\), \(p_{{_{2} }}\), and \(p_{3}\) denotes the random numbers. \(p_{1}\) parameter is balanced by exploration and exploitation, which is given in Eq. (14).
where b and B signifies the current and maximum iteration. \(p_{{_{2} }}\) and \(p_{3}\) are in the range [0, 1]. An objective function is calculated to evaluate the EXGB model’s performance with the set of hyperparameters in each iteration. Initially, the images are divided into two sets: the validating set and the training set. The objective function is determined in Eq. (15).
where \(k_{1}\) and \(k_{2}\) denotes the index sets of validating and training images, \(L_{1}\) and \(L_{2}\) signifies the \(k_{1}\) and \(k_{2}\) cardinality, \(r_{i} \,and\,\hat{r}\) signifies the predicted and actual value.The objective function combines the training and validation errors to avoid the problem of overfitting. The position of followers is updated utilizing Eq. (16).
where t and \(Z_{o}\) denotes the time and initial speed,\(E = \frac{{Z_{finl} }}{{Z_{o} }}\) represents the acceleration, and \(Z_{finl} = \frac{{X - X_{o} }}{t}\) with \(X_{o}\) and \(X\) represent final and initial locations.
The salps are initially still (\(Z_{o} = 0\)), hence the updated position of FS is signified utilizing Eq. (17).
Introducing the inertia weight parameter \((\beta \in (0,1))\) in the SSO enhances both exploitation and exploration capabilities, accelerating the convergence speed during the search process. The formulation for the ESSO is provided in Eqs. (18) and (19).
The process of updating the location of the sap is repeated until a predetermined number of iterations is reached or an optimal EXGB hyperparameter is found. Utilizing the optimized hyperparameters, the EXGB model demonstrates superior performance. Consequently, the CEXGB model exhibits precise detection and classification of brain tumors, encompassing Glioma Tumor, Pituitary Tumor, and Meningioma Tumor. Figure 2 illustrates the working procedure of the ESSO-EXGB model.
4 Experimental Results and Analysis
The experiment is conducted on ‘Brain Tumor Classification (MRI)’dataset [22].Brain tumor is divided into Benign Tumor, Pituitary Tumor, glioma, and Meningioma Tumor. There are105 images in Benign Tumor, 74 images in Pituitary Tumor, 115 images in Meningioma tumor, and 100 images in glioma tumor.
4.1 Simulation Setup
A brain tumor detection and classification model based on the CEXGB-ESSO model is evaluated in Python. All computations are done on Google Colab, which runs Ubuntu 64-bit and has a single-core hyper-threaded Intel Xeon processor clocked at 2.3 GHz and 13 GB RAM. The detection accuracy is improved by tuning the maximum depth-10, gamma-1, number of estimators-200, minimum child weight-5, and learning rate-0.3. The hyperparameters for ESS optimization are an inertia weight of 0.7 and an iteration of 200. Table 1 presents the CNN-EXGB settings.
4.2 Evaluation Metrics
The detection and classification efficiency are analyzed by some measures, such as sensitivity, precision, specificity, and prediction accuracy. The four parameters used to determine these performance measures are ‘TP’ (true positive), ‘TN’(true negative), ‘FP’(false positive),and ‘FN’(false negative).
(i) AccuracyIt measures the ratio of correct classification to overall classification.
(ii) Sensitivity It is the total number of positives that can be accurately determined.
(iii) Specificity It is the ratio of those who do not have tumor who test negatives, which is given in Eq. (22).
(iv) Precision It signifies the number of tumor correctly detected as positive from total identified as positive, which is given in Eq. (23).
4.3 Performance Evaluation
Here, the proposed CEXGB-ESSO model is comprehensively evaluated and compared with GResNet [15], RNIWO [17], DarkNet [18], SRELU [21], NASNet [24], and hybrid CNN-SVM [23]. The evaluation metrics encompass accuracy, precision, sensitivity, and specificity, providing a holistic assessment of the model’s performance across different tumor classes.
Figure 3 depicts the accuracy comparison between the suggested CEXGB-ESSO model and existing methods. Traditional methods, reliant on backpropagation in final layers, tend to escalate model complexity. In contrast, the CEXGB-ESSO model streamlines the process using CEXGB, significantly reducing calculation parameters. As a result, the proposed CEXGB-ESSO model achieves a remarkable accuracy of 99%, surpassing GResNet (94%), RNIWO (96%), DarkNet (96.8%), SRELU (97%), hybrid CNN-SVM (94%), and NASNet (95%) for glioma type brain tumors.
Figure 4 illustrates precision comparisons for different brain tumor classes. Traditional methods often struggle with parameter selection, leading to suboptimal performance. The proposed CEXGB-ESSO model employs ESSO for optimal hyperparameter tuning, significantly enhancing precision. Consequently, proposed CEXGB-ESSO model achieves high precision (98%) compared to GResNet (93%), RNIWO (95.5%), DarkNet (96.2%), SRELU (96.6%), hybrid CNN-SVM (94.3%), andNASNet (93.5%) for glioma type brain tumors.
Figure 5 showcases sensitivity comparisons, emphasizing the impact of ESSO optimization on detection accuracy. The inertia parameter introduced by ESSO enhances both exploration and exploitation capabilities, resulting in high sensitivity (96.5%) for the proposed model. This outperforms GResNet (93%), RNIWO (93.7%), DarkNet (93.8%), SRELU (94.4%), hybrid CNN-SVM (91.5%), and NASNet (93%) for glioma type brain tumors.
Figure 6 highlights specificity comparisons, emphasizing the efficacy of the CEXGB model in automatically extracting features. The convolution layers in CEXGB contribute to high specificity (97%) for the proposed CEXGB-ESSO model compared to GResNet (94%), RNIWO (92.9%), DarkNet (94.8%), SRELU (93.4%), NASNet (93.5%), and hybrid CNN-SVM (92%) for glioma type brain tumors.
Figure 7 provides the comparison of accuracy with the number of epochs for the proposed CEXGB-ESSO model with existing methods. At the beginning, accuracy is minimum for all method, as the epochs increase, our proposed method attains high accuracycompared to other methods. Our proposed CEXGB-ESSO model (90%) provides improved performance than the existing methods such as GResNet (78%), RNIWO (73%), DarkNet (89%), and SRELU (80%).Table 2 provides the values of the Figs. 4, 5, 6, And 7 in numericalvalue.
Table 3 provides the output obtained for detection and classification using the CEXGB-ESSO model. Out of some samples, one sample is selected and tabulated as the result for meningioma, pituitary, and glioma tumor. The detected and classified tumor region is shown in a red box. As shown in the table, the box accurately encloses the tumor-affected region of the MRI. This indicates that the detection system used is capable of detecting and classifying brain tumors
Further, a convergence analysis is conducted to showcase the effectiveness of ESSO in enhancing the convergence speed of the proposed CEXGB-ESSO model. ESSO is compared with existing optimization algorithms, namely Standard Salp Swarm Optimization (SSO), Firefly Algorithm (FA), Modified FA (mFA), and Whale Optimization Algorithm (WOA). These algorithms were chosen to represent distinct optimization paradigms and offer a comprehensive comparison. SSO serves as a baseline, while FA and its modified version (mFA) represent different swarm intelligence approaches. The inclusion of WOA adds diversity to the comparison. The convergence curves and results will be presented, providing insights into the relative performance and showcasing how ESSO excels in accelerating convergence for brain tumor detection and classification. Figure 8 illustrates the convergence performance of these algorithms. It is evident from the comparison that ESSO consistently exhibits a faster convergence rate, reaching optimal hyperparameter values approximately around iteration 25. In contrast, other algorithms, including SSO, FA, mFA, and WOA, demonstrate comparatively slower convergence. The convergence analysis emphasizes the efficiency of ESSO in finding optimal hyperparameter values within a shorter time frame. This enhanced convergence speed is crucial for the proposed CEXGB-ESSO model, as it contributes to the overall efficiency and accuracy in brain tumor detection and classification. The comparison provides insights into the relative performance of different optimization algorithms, with ESSO standing out as a favorable choice for accelerating convergence in the proposed model.
4.4 Ablation Analysis
In this section, an ablation study is carried out to evaluate the individual contributions of key components in the CEXGB- ESSO model. The experiments aim to elucidate the impact of each element on the overall performance of brain tumor classification.
4.4.1 Effectiveness of ESSO
To demonstrate the effectiveness of CEXGB, comparing it with a CNN without EXGB, and EXGB.
-
CNN without EXGB represent network without the EXGB component. This can help establish a baseline for the contribution of EXGB to the model’s effectiveness.
-
EXGB represent the model’s performance when using only EXGB without the CNN. This will help understand the role of EXGB in the absence of CNN features.
Table 4 shows that the CNN-only configuration achieved respectable performance but lacked the boosting effect inherent in the EXGB component. The standalone EXGB configuration demonstrated the algorithm’s capability for accurate predictions, serving as a baseline. Notably, the proposed CEXGB model, combining CNN and EXGB, outperformed both individual configurations, showcasing a synergistic effect. The integration of convolutional features and boosting techniques led to superior accuracy, sensitivity, precision, and specificity, affirming the efficacy of the suggested hybrid model. Also, the proposed CNN with EXGB (CEXGB has lower number of parameters than the standalone CNN without EXGB. This demonstrates the efficiency of the CEXGB model in minimizing parameters while preserving its effectiveness in brain tumor detection and classification. This detailed analysis provides valuable insights into the distinctive contributions of each model component, enhancing our understanding of the CEXGB model’s overall effectiveness
4.4.2 Effectiveness of ESSO
To demonstrate the effectiveness of ESSO, comparing it with a scenario without ESSO, with SSO but without an inertia weight parameter, and with ESSO:
-
Full Model represents the complete CEXGB-ESSO model.
-
Without ESSO represents the model without the Enriched Salp Swarm Optimization.
-
With SSO but without inertia represents the model with SSO but without the inertia weight parameter.
Table 5 provides the performance of the suggested CEXGB-ESSO for detection and classification of brain tumor, considering various configurations. The full model achieves the highest accuracy (99%) and sensitivity (97.525%), serving as the baseline. Removing ESSO results in a decrease in accuracy and sensitivity, emphasizing the significance of ESSO in enhancing overall performance. Utilizing Standard Salp Swarm Optimization without the inertia weight parameter leads to a further decline in accuracy and sensitivity. This ablation study underscores the novel contribution of ESSO, showcasing its impact on convergence speed and accuracy, making it a crucial component in the proposed brain tumor classification model
The superior performance of the proposed CEXGB-ESSO model can be attributed to its ability to leverage both convolutional features and boosting techniques. The integration of EXGB enhances the model’s capacity to detect and classify brain tumors accurately, as demonstrated in both the performance evaluation and ablation analyses. The ablation study further elucidates the novel contributions of ESSO, emphasizing its role in accelerating convergence and improving overall model performance. In conclusion, the proposed CEXGB-ESSO model stands out as an efficient and accurate approach for brain tumor detection and classification. The ablation study provides a nuanced understanding of the model’s components, shedding light on the unique contributions of CEXGB and ESSO.
5 Conclusion
In this research paper, CEXGB-ESSO model is introduced to detect and classify brain tumor. Initially, the MRI image is fed to bilateral filtering to remove noise. After that, the de-noised image is given to the CEXGB-ESSO model. In CNN, the fully connected layer is replaced by EXGB because it minimizes the number of parameters without back propagation. The integration of CNN with EXGB in the proposed model synergistically enhances feature extraction and classification accuracy. ESSO fine tunes the EXGB hyperparameters. This algorithm contributes to faster convergence, improving the efficiency of the model. Through a comprehensive evaluation against state-of-the-art techniques, including GResNet, RNIWO, DarkNet, SRELU, NASNet, and hybrid CNN-SVM, proposed CEXGB-ESSO model demonstrates superior performance across precision (98.2), accuracy (99), sensitivity (97.52), and specificity (97.7).This proposed CEXGB-ESSO model not only contributes to the field of medical image analysis but also underscores the significance of combining deep learning and optimization techniques for improved diagnostic capabilities in healthcare. However, the tumor substructure like necrotic and solid cannot be find. So in the future, a model is designed to identify the tumor substructures.
References
Reddy S, Tatiparti K, Sau S, Iyer AK (2021) Recent advances in nano delivery systems for blood-brain barrier (BBB) penetration and targeting of brain tumors. Drug Discovery Today 26(8):1944–1952
Polat Ö, Güngen C (2021) Classification of brain tumors from MR images using deep transfer learning. J Supercomput 77(7):7236–7252
Rivera M, Norman S, Sehgal R, Juthani R (2021) Updates on surgical management and advances for brain tumors. Curr Oncol Rep 23(3):1–9
Khan AR, Khan S, Harouni M, Abbasi R, Iqbal S, Mehmood Z (2021) Brain tumor segmentation using K-means clustering and deep learning with synthetic data augmentation for classification. Microsc Res Tech 84(7):1389–1399
Paul J, Sivarani TS (2021) Computer aided diagnosis of brain tumor using novel classification techniques. J Ambient Intell Humaniz Comput 12(7):7499–7509
Mzoughi H, Njeh I, Slima MB, Ben Hamida A, Mhiri C, Mahfoudh KB (2021) Towards a computer aided diagnosis (CAD) for brain MRI glioblastomas tumor exploration based on a deep convolutional neuronal networks (D-CNN) architectures. Multimed Tools Appl 80(1):899–919
Worrell SL, Kirschner ML, Shatz RS, Sengupta S, Erickson MG (2021) Interdisciplinary approaches to survivorship with a focus on the low-grade and benign brain tumor populations. Curr Oncol Rep 23(2):1–8
Elshaikh BG, Garelnabi ME, Omer H, Sulieman A, Habeeballa B, Tabeidi RA (2021) Recognition of brain tumors in MRI images using texture analysis. Saudi J Biol Sci 28(4):2381–2387
Badrigilan S, Nabavi S, Abin AA, Rostampour N, Abedi I, Shirvani A, Ebrahimi Moghaddam M (2021) Deep learning approaches for automated classification and segmentation of head and neck cancers and brain tumors in magnetic resonance images: a meta-analysis study. Int J Comput Assist Radiol Surg 16(4):529–542
Overcast WB, Davis KM, Ho CY, Hutchins GD, Green MA, Graner BD, Veronesi MC (2021) Advanced imaging techniques for neuro-oncologic tumor diagnosis, with an emphasis on PET-MRI imaging of malignant brain tumors. Curr Oncol Rep 23(3):1–5
Biswas A, Islam MS (2021) Brain tumor types classification using k-means clustering and ANN approach. In: 2021 2nd international conference on robotics, electrical and signal processing techniques (ICREST) IEEE, 654–658
Deepak S, Ameer PM (2021) Automated categorization of brain tumor from mri using cnn features and svm. J Ambient Intell Humaniz Comput 12(8):8357–8369
Kumar DM, Satyanarayana D, Prasad MN (2021) MRI brain tumor detection using optimal possibilistic fuzzy C-means clustering algorithm and adaptive k-nearest neighbor classifier. J Ambient Intell Hum Comput. 2867–280.
Chen B, Zhang L, Chen H, Liang K, Chen X (2021) A novel extended kalman filter with support vector machine based method for the automatic diagnosis and segmentation of brain tumors. Comput Methods Programs Biomed 200:105797
Kumar RL, Kakarla J, Isunuri BV, Singh M (2021) Multi-class brain tumor classification using residual network and global average pooling. Multimed Tools Appl 80(9):13429–13438
Majib MS, Rahman MM, Sazzad TS, Khan NI, Dey SK (2021) Vgg-scnet: a vgg net-based deep learning framework for brain tumor detection on mri images. IEEE Access 9:116942–116952
Dixit A, Nanda A (2021) An improved whale optimization algorithm-based radial neural network for multi-grade brain tumor classification. Vis Comput. 1–6
Ahuja S, Panigrahi BK, Gandhi TK (2022) Enhanced performance of Dark-Nets for brain tumor classification and segmentation using colormap-based superpixel techniques. Mach Learn Appl 7:100212
Sharif MI, Khan MA, Alhussein M, Aurangzeb K, Raza M (2021) A decision support system for multimodal brain tumor classification using deep learning. Complex Intell Syst. 1–4
Sasank VV, Venkateswarlu S (2021) Brain tumor classification using modified kernel based soft plus extreme learning machine. Multimed Tools Appl 80(9):13513–13534
Alhassan AM, Zainon WM (2021) Brain tumor classification in magnetic resonance image using hard swish-based RELU activation function-convolutional neural network. Neural Comput Appl. 1–3
Bhuvaji S, Kadam A, Bhumkar P, Dedge S, Kanchan S (2020) Brain tumor classification (MRI). Kaggle. https://doi.org/10.34740/KAGGLE/DSV/1183165
Khairandish MO, Sharma M, Jain V, Chatterjee JM, Jhanjhi NZ (2022) A hybrid CNN-SVM threshold segmentation approach for tumor detection and classification of MRI brain images. Irbm 43(4):290–299
Sadad T, Rehman A, Munir A, Saba T, Tariq U, Ayesha N, Abbasi R (2021) Brain tumor detection and multi-classification using advanced deep learning techniques. Microsc Res Tech 84(6):1296–1308
Nawaz SA, Khan DM, Qadri S (2022) Brain tumor classification based on hybrid optimized multi-features analysis using magnetic resonance imaging dataset. Appl Artif Intell 36(1):2031824
Kesav N, Jibukumar MG (2022) Efficient and low complex architecture for detection and classification of brain tumor using RCNN with two channel CNN. J King Saud Univ Comput Inf Sci 34(8):6229–6242
Funding
There is no funding for this study.
Author information
Authors and Affiliations
Contributions
All the authors have participated in writing the manuscript and have revised the final version. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors declares that they have no conflict of interest.
Ethical Approval
This article does not contain any studies with human participants and/or animals performed by any of the authors.
Informed Consent
There is no informed consent for this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jebastine, J. Detection and Classification of Brain Tumor Using Convolution Extreme Gradient Boosting Model and an Enhanced Salp Swarm Optimization. Neural Process Lett 56, 135 (2024). https://doi.org/10.1007/s11063-024-11590-4
Accepted:
Published:
DOI: https://doi.org/10.1007/s11063-024-11590-4